By Aitor Balmaseda*, Nicolas Rozès, Albert Bordons and Cristina Reguant
Sourced from: Balmaseda, A., Rozès, N., Bordons, A., Reguant, C. (2021). Simulated lees of different yeast species modify the performance of malolactic fermentation by Oenococcus oeni in wine-like medium. Food Microbiology 99, 103839. https://doi.org/10.1016/j.fm.2021.103839
Oenococcus oeni is the main agent of wine malolactic fermentation (MLF) [1]. This fermentation is desirable in red wines and high acidity wines and usually takes place after the alcoholic fermentation (AF) that produces wine from grape must. During this process, oenological yeasts transform the sugars from the must into ethanol and CO2, while other nutrients are also exhausted. Traditionally to better control the AF, Saccharomyces cerevisiae is inoculated in the initial must. Nevertheless, during the last few decades, the increasing interest about the positive influence of non-Saccharomyces in wine opened a new scenario of metabolic activities and wine modulation in the final product [2]. This vast group of yeasts named non-Saccharomyces include other genera as Torulaspora, Metschnikowia, Lachancea, Hanseniaspora, Candida, etc.
Wine is a complex matrix with low nutrient content and high concentration of inhibitor compounds, which will be very dependent on the fermenting yeast metabolic activities during AF [3]. Besides, it is the preferent ecological niche of O. oeni where it has to grow and develop. In this sense, MLF is a stress metabolism that enables the bacterium to survive with a not very efficient energy source: L-malic acid [4].
Under these harsh conditions, the failure of MLF can occur due to the low nutrient content and high inhibitory compounds present in wine. That is why in some cases, nutrient supplementation is used as a strategy to promote MLF. Indeed, MLF is usually performed in presence of yeast lees under cellar conditions. The lees, mainly deposits of dead yeast or residual yeast, eventually suffer an autolytic process that contribute to increase nutrient concentration in wine. In this sense, the contribution of yeast autolysates to wine characteristics can be affected by yeast species.
In this study [5], we tested the MLF performance of three O. oeni strains in wine-like medium (WLM) in presence of yeast lees of different strains belonging to S. cerevisiae (ScQA23, Sc3D), T. delbrueckii (TdBiodiva, TdViniferm, TdTDP) and M. pulcherrima (MpFlavia, MpMPP) species. These lees were obtained by growing the yeasts in natural grape must. Then, they were inactivated by heat and mechanical treatment.
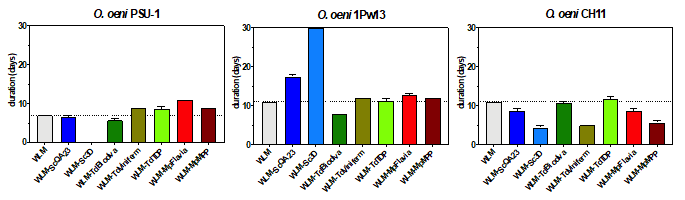
We observed that the addition of yeast lees had different impact on MLF duration depending on the O. oeni strain. In general, yeast lees had a positive effect on strain CH11, a neutral effect on strainPSU-1, and a negative effect onstrain 1Pw13 (Figure 1). In this sense, the addition of non-Saccharomyces lees was more likely related to a stimulatory effect. Contrary, S. cerevisiae lees caused a great delay in MLF of O. oeni 1Pw13, and also a stuck fermentation in O. oeni PSU-1 fermenting in WLM with S. cerevisiae 3D lees.
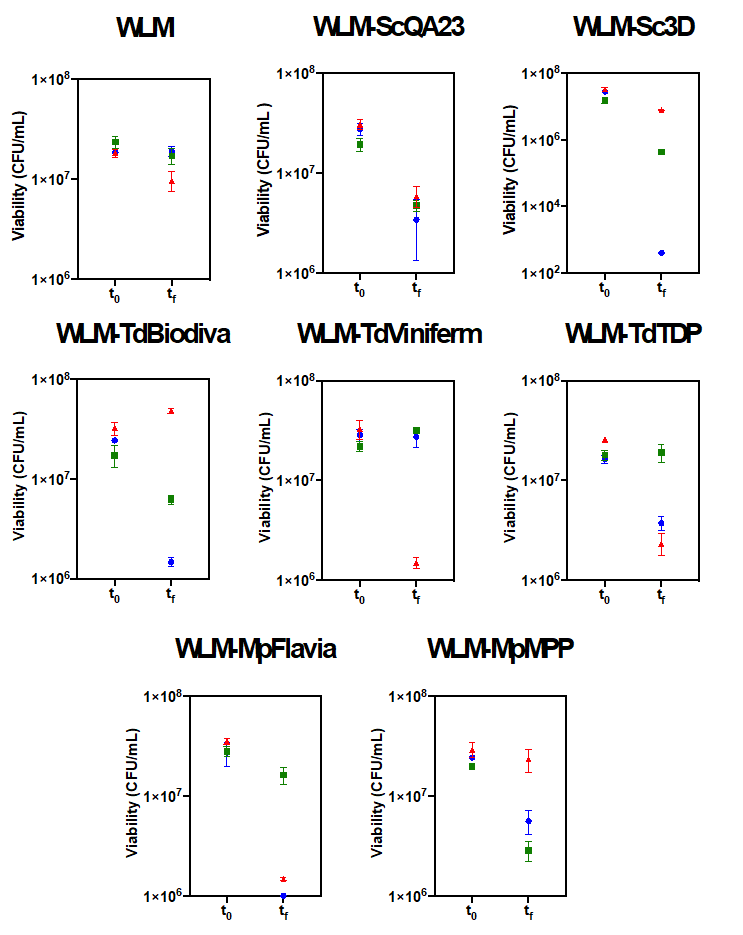
There was also a significant effect of these yeast lees in the viability of O. oeni (Figure 2). There was a general decrease in viability that depended on the added lees and the inoculated O. oeni strain. O. oeni PSU-1 was the strain that lost the most viability in all wines. Indeed, this strain suffered a dramatic viability decrease in WLM-Sc3D. On the contrary, viability in WLM-TdViniferm was maintained pointing a positive effect of this yeast on O. oeni viability. The other two O. oeni strains generally maintained their viability with some exceptions where it was increased, always with non-Saccharomyces lees.
We evaluated also other compounds and metabolisms important in MLF as citric acid consumption or mannoprotein utilisation that were significantly affected by the use of non-Saccharomyces [5]. Overall, we observed that non-Saccharomyces lees presented a more MLF friendly environment for O. oeni which was generally related with better MLF performances. Nevertheless, this effect was different depending on the fermenting O. oeni strain with points a complex interactions patterns between yeast lees – O. oeni compatibility.
To sum up, the use of non-Saccharomyces lees opens a new scenario to produce oenological products based in these yeasts in the development of stimulating MLF extracts.

Universitat Rovira i Virgili, Departament de Bioquímica i Biotecnologia, Facultat d’Enologia, 1Grup de Biotecnologia Microbiana dels Aliments, 2Grup de Biotecnologia Enològica, C/ Marcel·lí Domingo s/n, 43007 Tarragona, Catalonia, Spain
Both research groups (1 & 2) focus mainly their studies on the role of wine microorganisms in wine quality. They collaborate with the aim of studying the interaction between yeast and lactic acid bacteria in wine and its impact on malolactic fermentation.
References
- Lonvaud-Funel A. (1999) Lactic acid bacteria in the quality improvement and depreciation of wine. In: Konings W.N., Kuipers O.P., In ’t Veld J.H.J.H. (eds) Lactic Acid Bacteria: Genetics, Metabolism and Applications. Springer, Dordrecht. https://doi.org/10.1007/978-94-017-2027-4_16
- Padilla, B., Gil, J.V., Manzanares, P. (2016). Past and future of non-Saccharomyces yeasts: from spoilage microorganisms to biotechnological tools for improving wine aroma complexity. Front. Microbiol. 7 https://doi.org/10.3389/fmicb.2016.00411.
- Balmaseda, A., Bordons, A., Reguant, C., Bautista-Gallego, J. (2018). Non-Saccharomyces in wine: effect upon Oenococcus oeni and malolactic fermentation. Front. Microbiol. 9, 534. https://doi.org/10.3389/fmicb.2018.00534.
- Davis, C.R., Wibowo, D., Eschenbruch, R., Lee, T.H., Fleet, G.H. (1985). Practical implications of malolactic fermentation: a review. Am. J. Enol. Vitic. 36, 290–301.
- Balmaseda, A., Rozès, N., Bordons, A., Reguant, C. (2021). Simulated lees of different yeast species modify the performance of malolactic fermentation by Oenococcus oeni in wine-like medium. Food Microbiol. 99, 103839. https://doi.org/10.1016/j.fm.2021.103839.

