By Robert Powers, Eric V. Anslyn, Ron C. Runnebaum and Diana Zamora-Olivares
The adulteration, counterfeiting or relabeling of wine is a growing world-wide problem due to its economic and cultural value. Thus, obtaining a chemical fingerprint for a wine to validate its vintage, vineyard or region of origin has been extensively studied. Both biological and chemical analytical techniques have been used to acquire a chemical or metabolomic profile (i.e., identification and quantification of the complete set of small molecules within the wine sample). These chemical profiling techniques vary from sequencing technologies1, 2 to nuclear magnetic resonance (NMR) spectroscopy.3-5 Metabolomics studies have used a combination of quantitative untargeted NMR data with chemometrics to distinguish wines based on grape varieties,6 region of growth,6, 7 effects of vintage year,4, 8 and vinification approach.9
In addition to traditional analytical techniques, the differential sensing (DS) approach along with chemometric routines10, 11 has become a powerful alternative method to detect and distinguish between a variety of complex biological samples.12, 13 For wine, the DS technique mimics the senses of taste, smell, and palate texture by using a variety of cross-reactive receptors that display different binding affinities for multiple target molecules.14 The DS method has been successfully employed to classify wine varietals,15 wine blends,16 and to differentiate harvest decisions17 on the basis of phenolic composition and distribution. A novel strategy was explored by combining an efficient and relatively simple readout of targeted DS arrays with a complementary untargeted approach offered by one-dimensional (1D) 1H NMR spectroscopy. The combination of both analytical methods improved the analysis outcome and classification of wine providing results that neither method can efficiently achieve independently (Figure 1). This combinatorial strategy was crucial to capture subtle variations uniquely imparted by growing site and year to year growing conditions. Thus, we hypothesized that the combination of these two analytical techniques would improve the classification of Pinot noir wines grown from fifteen vineyards, from eight distinct American Viticultural Areas (AVAs) along the United States Pacific Coast, and for two vintage years (2015 and 2016).
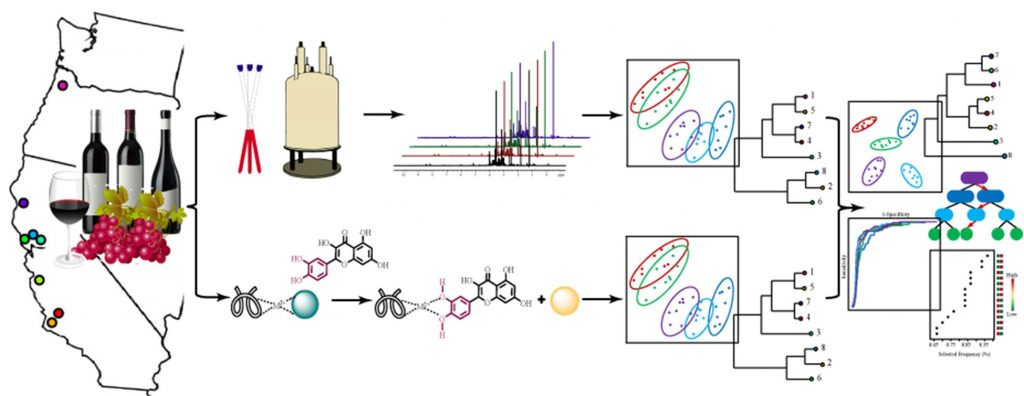
The Pinot noir wines were classified with a very high accuracy according to vineyard, regions, and vintage by combining 1D 1H-NMR spectroscopy with DS arrays. This was achieved by using multivariate PCA models and univariate statistical modeling based on Random Forest (RF) and receiver operating characteristic (ROC) curve analyses. Variability attributed to fermentation and aging steps were minimized by using identical fermentation and aging protocols in stainless steel vessels. Quantifying differences that could be attributed to the growing site or conditions was of upmost importance considering the anticipated changes in microclimates and water availability over the upcoming decades due to climate change.18

ROC curves were used to identify DS array and NMR spectroscopy features that were frequently utilized to distinguish Pinot noir wines by vineyard or AVA region (Figure 2). The top features selected from the ROC curve analysis of the combined NMR spectroscopy and DS array dataset for the 2015 SNC2 vineyard are shown in Figure 2A. The ROC feature selection plot in Figure 2A identifies the specific NMR bins (i.e., ppm) and peptide sensors (i.e., MM1–MM9) that differentiate the 2015 SNC2 vineyard from the remaining vineyards. The plot also identifies the relative directional change in the features and how often each feature was selected to differentiated between the vineyards. A similar analysis was completed for each AVA region, where a representative plot of the top features is shown in Figure 2B for the 2015 SNC AVA region. The top selected features from all ROC curve analyses were cataloged and the usage-frequency between vineyards and AVA regions are plotted in Figures 2C and 2D, respectively. The frequency plots identified the NMR spectroscopy features that were frequently selected as discriminators of the Pinot noir wines. These features were then assigned to potential metabolites or classes using a set of reference 1D 1H NMR spectra of known wine metabolites. Specifically, 55 metabolites were previously identified from four prior NMR metabolomics studies of similar wines.6, 19-21 Figure 2C and 2D depict several metabolite classes and putative metabolite IDs that may influence Pinot noir wines according to vineyard or AVA region. Our analysis suggests that metabolites such as branched-chain amino acids, sugar alcohols including ethyl alcohols and phenyl alcohols, tricarboxylic acid cycle derivatives such as malic acid and citric acid, sugars including fructose, and aromatic amino acids may play an important role in classifying Pinot noir wines across the Pacific Coast of the United States. It is important to note that the primary purpose of our study was to demonstrate the value of combining NMR and DS array features to classify and differentiate Pinot noir wines based on vineyard, region, and vintage. Conversely, assigning a metabolite to each of the differentiating features was not an original intent. The ROC curves further supported the observation that the 2015 and 2016 vintage years exhibited distinct features that were independent of vineyard and AVA region. The univariate analyses (RF and ROC) clearly highlighted the value and importance of combining multiple analytical techniques to identify distinct regions of the metabolic fingerprint. In addition to the improved accuracy of predicting vineyard or AVA region membership, the univariate analyses corroborate that different combinations of features were required to accurately classify each vineyard or AVA region. Along with vineyard and AVA region classification, vintage year was also evaluated for a metabolic fingerprint that contributed to distinguishing the various Pinot noir wines. As our results indicate, we have found that the AVA region, and more importantly, the specific vineyard site, can significantly impact the metabolic fingerprint of the Pinot noir wines.
Herein, we report on the combination of multiple analytical techniques to improve the classification accuracy of identical Pinot noir clones grown across distinct geographic locations. Toward this end, we report that the combination of untargeted metabolomic fingerprinting using 1D 1H NMR spectroscopy with a targeted analysis of phenolic profiles using a colorimetric DS peptide-based array has proven to be a highly effective approach to distinguish wines produced from genetically identical grapevines across vineyard location, geographic region, and vintage year. Our analysis highlights that targeted and untargeted techniques can be combined to successfully classify wine varietals solely based on geographic location and vintage year.
Reference Footnote
Reprinted from Food Chemistry, 354, Crook, A. A.; Zamora-Olivares, D.; Bhinderwala, F.; Woods, J.; Winkler, M.; Rivera, S.; Shannon, C. E.; Wagner, H. R.; Zhuang, D. L.; Lynch, J. E.; Berryhill, N. R.; Runnebaum, R. C.; Anslyn, E. V.; Powers, R., Combination of two analytical techniques improves wine classification by Vineyard, Region, and vintage, 129531, Copyright (2021), with permission from Elsevier. Detailed information can be found at https://doi.org/10.1016/j.foodchem.2021.129531

Robert Powers 
Eric V. Anslyn 
Ron Runnebaum 
Diana Zamora-Olivares
Robert Powers – Dr. Robert Powers is a Professor of Chemistry at the University of Nebraska-Lincoln, Director of the Systems Biology core facility within the Nebraska Center for Integrated Biomolecular Communication, and is on the scientific advisory board for Olaris Therapeutics, Inc and Nexomics Biosciences, Inc. Dr. Powers received his BA from Rutgers University, a Ph.D. in chemistry from Purdue University, and was an IRTA postdoctoral fellow at NIH. Prior to UNL, Dr. Powers was the Associate Director for the Protein NMR group at the pharmaceutical company, Wyeth. Dr. Powers is the founding Editor-in-Chief of Current Metabolomics, is a board member of the Metabolomics Association of North America, a member of the Metabolomics Quality Assurance & Quality Control Consortium, is on the Editorial Board of Nature Scientific Reports and 6 other journals, is an AAAS fellow, and recipient of the ACS Outreach Volunteer of the Year and an ACS Chemistry Ambassador. He has written over 150 peer-reviewed manuscripts, 7 book chapters, is an inventor on 10 patents, and has given over 160 invited lectures.
Eric V. Anslyn – Dr. Eric Anslyn is the Welch Regents Chair of Chemistry at the University of Texas at Austin. He attended the California State University Northridge to receive his BS degree in Chemistry. After that he performed doctoral research at the California Institute of Technology under the direction of Dr. Robert Grubbs studying fundamental mechanisms of olefin metathesis. Subsequently, he was a National Science Foundation post-doctoral Fellow at Columbia University working with Dr. Ronald Breslow studying artificial enzymes. Dr. Anslyn has published over 350 papers, holds approximately 50 patents, and has given over 400 invited lectures. He has been recognized with a variety of research and teaching awards, including: The Centenary Prize from the RSC, The James Flack Norris Award from the ACS, the Izatt-Christensen Award, The Czarnik Award, the ACS Edward Leete Award, and the Arthur C. Cope Scholar Award from the ACS. For his novel advances in teaching, he was recently named an HHMI Professor, and he has won the Texas state-wide Regents Teaching Award.
Ron C. Runnebaum – Dr. Ron Runnebaum works on improving processes for more sustainable use of natural resources, including those important in winemaking. He is a faculty member in the Departments of Viticulture & Enology and Chemical Engineering at the University of California at Davis (UC Davis). His research program in the Department of Viticulture & Enology aims to combine his interests in sustainable winemaking with his research background in nanomaterials, adsorption, heterogeneous catalysis, and reaction engineering. Dr. Ron Runnebaum’s wine research includes elucidating contributions of vineyard site on wine chemistry/sensory and seeking alternatives for removing potassium bitartrate and proteins
Diana Zamora-Olivares – Dr. Diana Zamora-Olivares is an Assistant Professor at the University of Texas in Austin, and Associate Director of the UT Wine Initiative leading collaborative efforts with Central Texas vineyards. Dr. Zamora-Olivares attended the National Autonomous University of Mexico to receive her BS degree in Pharmaceutical Chemistry. She obtained her doctoral degree at the University of Texas at Austin under the direction of Professor Eric V. Anslyn developing differential sensing methods to detect, classify and quantify MAPK activity related to early-onset cancer. She was a NIH-Institutional Research and Academic Career Development Awards (IRACDA) post-doctoral fellow. Currently, she leads the FRI-Supra Sensors Chemistry Lab, which is an extension of the Anslyn Research Group.

