By Luciano Calderón and Diego Lijavetzky
‘Malbec’ is the signature grapevine (Vitis vinifera L.) cultivar of Argentina, mainly due to the international prestige reached by ‘Malbec’ wines elaborated in this country. Moreover, Mendoza is the region producing the greatest ‘Malbec’ wine volumes worldwide [1]. As most of the standout grape cultivars from other regions in the “new world” (like ‘Carménère’ in Chile or ‘Cabernet Sauvignon’ in California), ‘Malbec’ was also originated in France, more precisely in the Cahors region, during medieval times. It was later introduced to Mendoza during the 1850s, by the French agronomist Michel Aimé Pouget [2].
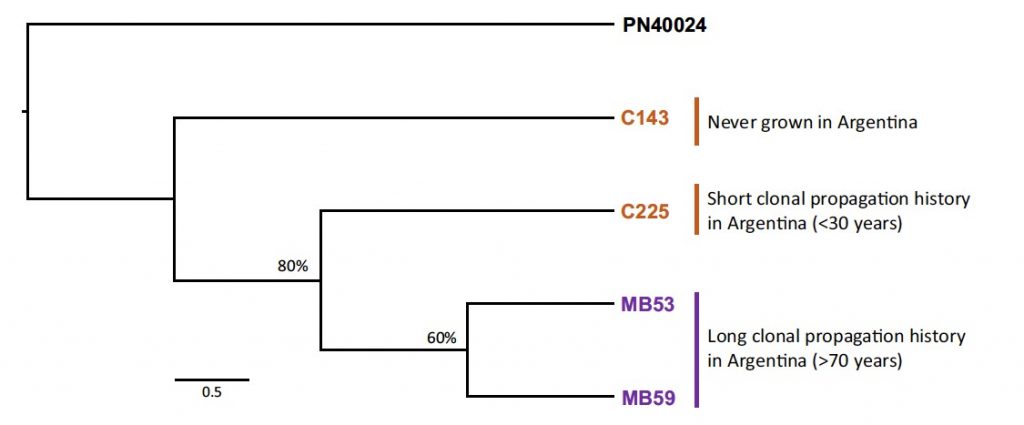
Previous works in ‘Malbec’ by our research group, reported a notorious clonal phenotypic diversity for different traits like berry biochemical composition or plant phenological development [3,4]. However, little was known about the clonal genetic diversity in this cultivar until now. That fact encouraged us to face the work recently published in Scientific Reports (doi.org/10.1038/s41598-021-87445-y), by analyzing the ‘natural’ clonal genetic diversity in ‘Malbec’. In that aim, we produced whole genome resequencing data for four clones with different clonal propagation records and origins. More precisely, two clones had a long history of clonal propagation in Argentina (>70 years), one clone was recently introduced from France (<30 years) and the other was never cultivated in Argentina. The phylogenetic analysis of this genomic data showed that the genetic relations among the analyzed accessions were associated to their clonal propagation records (Figure 1).
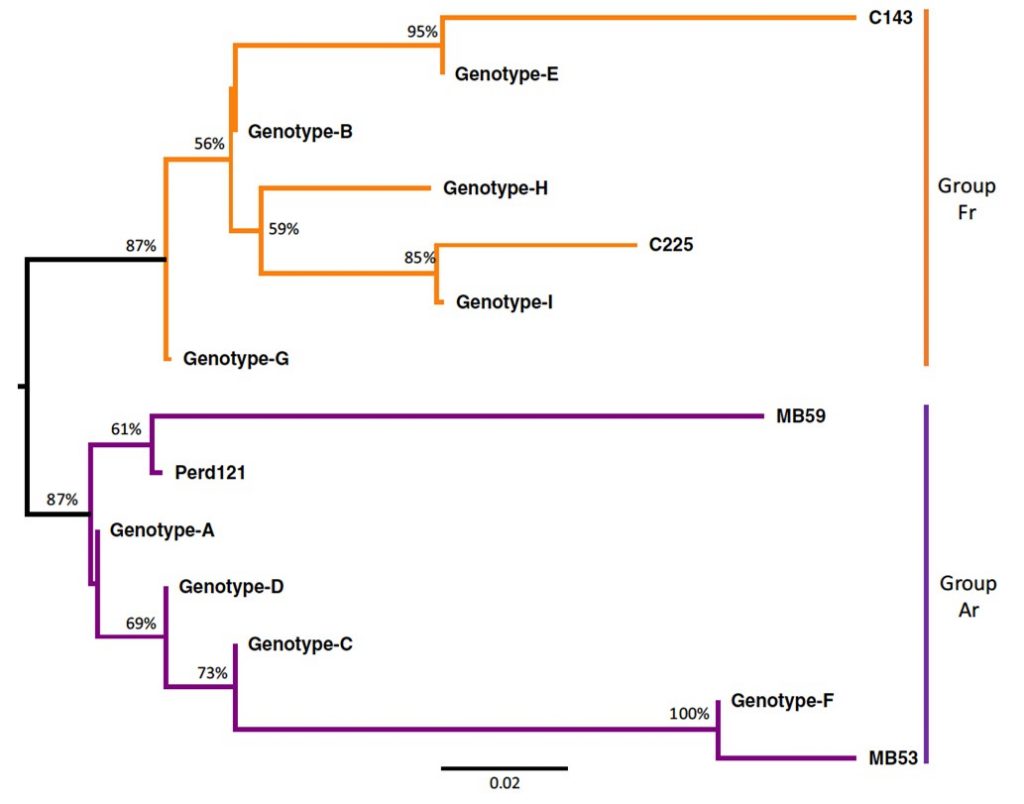
The obtained genomic data was also employed to identify a reliable set of genetic markers (single nucleotide variants), able to distinguish among clones. These markers were used to custom design a high-throughput Fluidigm genotyping chip (https://www.fluidigm.com/), employed to screen a wide sampling of ‘Malbec’ accessions obtained from, Vivero Mercier (Argentina), INTA EEA Mendoza (Argentina) and Finca El Encin (Spain). This experiment retrieved 14 different genotypes across the 214 analyzed accessions. Moreover, we observed that these genotypes clustered in two divergent clonal lineages. Group Ar included genotypes with a long history of clonal propagation in Argentina, and with a probable closer connection with the first ‘Malbec’ plants introduced to Mendoza during the 1850s. On the other hand, Group Fr included genotypes recently introduced or with a null history of clonal propagation in Argentina, with a more recent connection to the French origin of ‘Malbec’ (Figure 2).
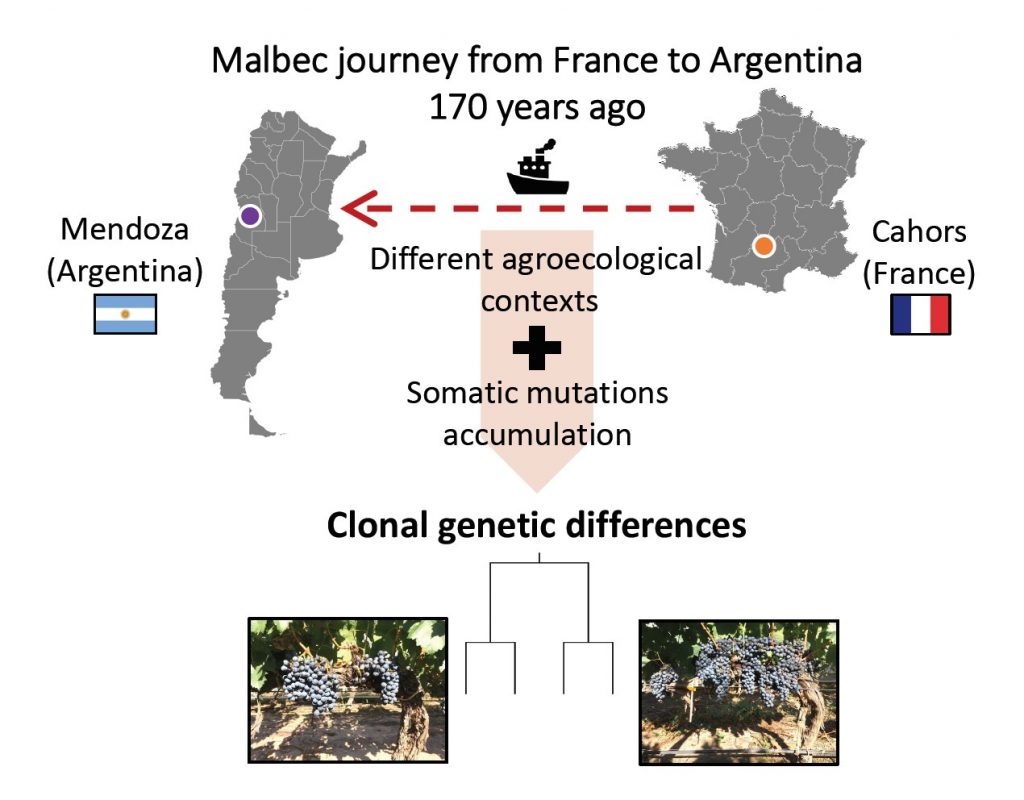
We conclude that the identified pattern of genetic diversity is likely the consequence of the combined effect of anthropic and natural biological processes. Plants transportation (from France to Argentina) and clonal selection, might have driven the accumulation of different somatic mutations, by exposing plants to different agroecological pressures on both sides of the Atlantic Ocean.
As final remarks, the developed genotyping chip could turn into a valuable tool for the wine industry, particularly for nurseries and producers. Custom genotyping for clone identification has already proved a useful tool to better understand clone’s history in ‘Chardonnay’ [5] and to trace clone geographic origin in ‘Nebbiolo’ [6]. Until now, with the available SSRs markers and SNPs chips, it was not possible to genetically distinguish among ‘Malbec’ clones. Here, we took advantage of high-throughput genomics and genotyping not only to provide insight on Malbec genetic diversity and history, but also to make available an objective method that could turn useful to improve clonal traceability. This work was performed by an international consortium of academic institutions: Instituto de Biología Agrícola de Mendoza (CONICET-UNCuyo) (Argentina), Laboratory of Virology, INTA-Mendoza (Argentina), Instituto de Ciencias de la Vid y del Vino (Spain) and Max Planck Institute for Developmental Biology (Germany); in collaboration with the nursery Vivero Mercier (Argentina).
Read more at: https://rdcu.be/cjPrP

Luciano Calderón 
Diego Lijavetzky 
Luciano Calderón, obtained his PhD degree in Biological Sciences at the University of Buenos Aires (2013). He performed research activities in genetics at several institutions worldwide (Canada, USA, Germany, Spain, Austria and Italy). Since 2019, he is a Junior Researcher for the Research Council of Argentina (CONICET), at the Group of Grapevine Genetics & Genomics (IBAM). Currently focused in understanding the genomic and transcriptomic mechanisms underlaying the intra-cultivar diversity in grapevines. Email contact: lucianocalderon@yahoo.com.ar
Diego Lijavetzky. Agronomist Engineer and PhD in Biological Sciences by the University of Buenos Aires. Postdoc at UC Davis and at the National Center for Biotechnology (CSIC, Madrid), where he started working in grapevine genetics and genomics. Since 2009, Senior Researcher at the National Research Council of Argentina (CONICET) and PI of the Group of Grapevine Genetics & Genomics (IBAM). The actual main projects are related to molecular studies regarding grapevine adaptation to climate change and grapevine transformation and gene editing by CRISPR-Cas9. https://www.researchgate.net/profile/Diego-Lijavetzky