by Javier Tello and Javier Ibáñez
Traditional viticulture systems are strongly threatened by climate change conditions, which compromise one of the most relevant agriculture sectors in Southern Europe. The selection (or generation) and use of more resilient grapevine genetic resources is suggested as one of the most efficient approaches to reach sustainable grape and wine production in climate-sensitive regions. This process requires of the previous characterization of the phenotypic diversity existing in the Vitis vinifera L. germplasm. Combined with genetic data, this information can be used to identify the genetic determinism of traits playing a role in the adaptation to climate change.
The genetic architecture of a trait has been traditionally revealed by the genetic study of segregating progenies. This approach is based on the generation of novel genetic variation by crossing two parents, generating an offspring of individuals that segregate for the trait of interest. This phenotypic variation, measured by quantitative systems, is combined to genetic information (obtained by the use of molecular markers) to detect the relationship between genetic and phenotypic variations. If a significant relationship is found, the surrounding genomic region is called “a quantitative trait locus”, or QTL.
That information can be obtained from the study of a large collection of grapevine genotypes too, in an approach termed linkage disequilibrium-based association mapping (AM). Whilst the idea of AM is rather similar as that of QTL detection in grapevine segregating progenies, it searches for functional associations in a broader context, using an association panel of grapevine cultivars selected by carrying most of the phenotypic diversity available at a species level for the trait of interest. This diversity derives from numerous historical and evolutionary recombination events happened across generations, which increases mapping resolution. Therefore, AM is of special interest for species with long generation cycles like the grapevine, as it avoids the laborious and time-cost process of establishing a mapping progeny. Given these advantages, AM is now the method of reference for many laboratories worldwide.
The experimental workflow of an AM study is shown in Figure 1. It includes the selection of the grapevine cultivars from the available germplasm, the evaluation of the trait(s) of interest (ideally in different locations during several seasons to overcome environmental fluctuations), and the screening of genetic markers. For a candidate gene association study (termed CGAS), these markers are genotyped at a locus (usually, one candidate gene) thought to be involved in the target trait. So, this approach requires of some previous knowledge of the genetic architecture of the trait. For a genome-wide association study (known as GWAS), markers are randomly obtained across the whole genome, through efficient genotyping technologies like genotyping-by-sequencing (GBS). Therefore, a GWAS is considered an exploratory approach. In both approaches, marker-trait association tests are conducted after genetic and phenotypic data collection, usually in software packages that integrate efficient modelling solutions that correct for population structure and kinship confounding effects. After the identification of significant statistical associations between genetic and phenotypic variations, these links have to be validated by independent approaches, as the risk of false positive associations is still present even under strong statistical evidence [1].
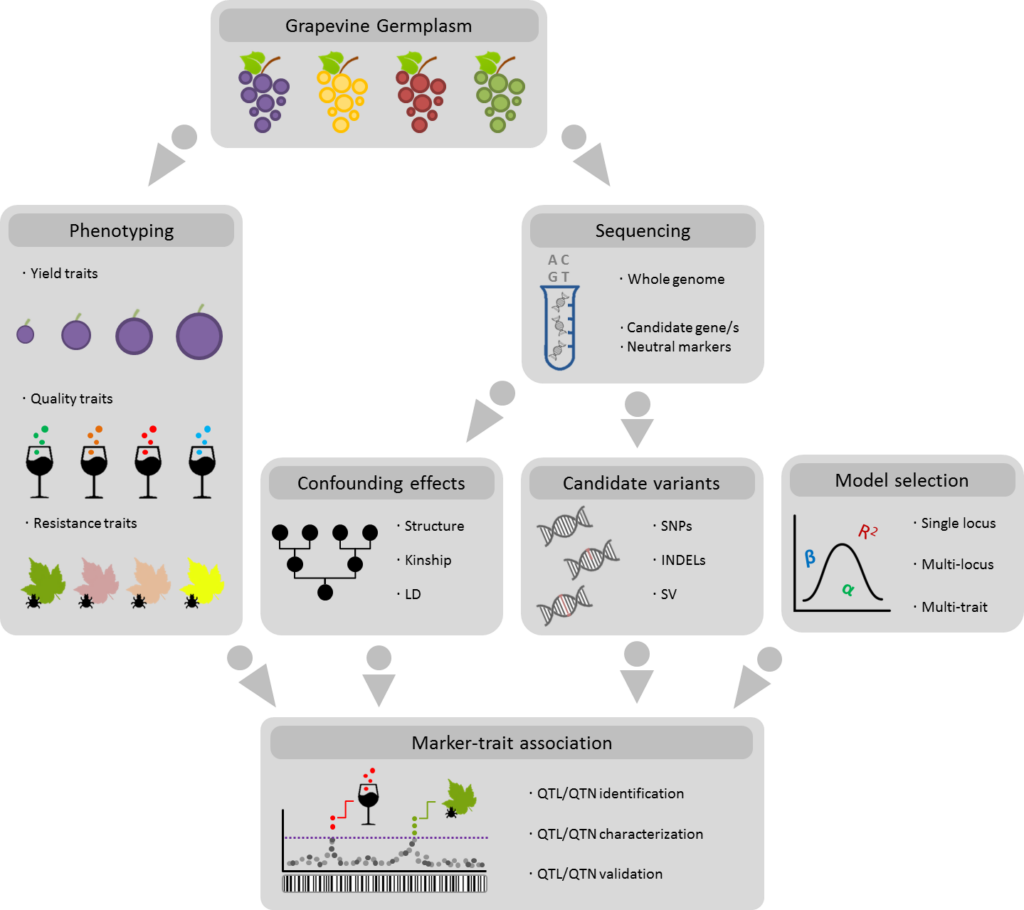
Figure 1. General approach to conduct an association mapping study in grapevine.
AM mapping has been used in grapevine research since 2007, when the first works exploring the association between phenotypic and genetic diversity were published. GWAS were reported later, when high-throughput sequencing technologies and new computational resources became available and cost-affordable. Nowadays, we can find AM studies exploring the genetic architecture of traits related to yield components, fruit quality, and resistance to biotic and abiotic factors.
Within the plethora of studies providing statistical relationship between allelic and phenotypic variations via AM, it is worthy to highlight the success of those exploring the genetic basis of berry colour, muscat aroma, and seedlessness.
Berry colour: berry colour has high commercial relevance for both wine and table grapes. Early genetic studies in segregating progenies indicated that this trait is mostly controlled by a single locus on chromosome 2, soon narrowed to a 200-kb region that clusters a series of VviMybA transcription factor genes. AM was useful to support the presence of a retrotransposon (Gret1) in the promoter region of VviMybA1 as the main causative element disrupting anthocyanins biosynthesis, whose presence leads to non-colored berries in V. vinifera cultivars. Additional polymorphisms in the VviMybA2 gene were also identified by AM as regulators of berry colour [2].
Muscat aroma: muscat varieties have been (and are) greatly appreciated for table and wine grape production. Multiple genetic studies in biparental progenies revealed a QTL of major effect on chromosome 5, which co-localizes with a 1-deoxy-D-xylulose 5-phosphate synthase (VviDXS1) gene that acts upstream in the biosynthesis of aromatic monoterpenoids. The analysis of this candidate gene by AM identified a SNP causing a non-neutral amino acid substitution in VviDXS1, which affects protein kinetics and increases monoterpenoids levels in muscat cultivars [3].
Seedlessness: stenospermocarpic seedlessness is one of the most prized traits in table grapes, as it produces berries of a relevant size without seeds. Genetic studies in either seedless × seedless or seeded × seedless biparental progenies found a major causative locus on chromosome 18, named SdI. Further analyses pointed out the grapevine AGAMOUS-LIKE11 (VviAGL11) gene as the functional candidate underlying SdI. Later on, AM studies identified an Arg-197Leu missense substitution as the functional mutation responsible for stenospermocarpic seedlessness [4].
These AM-derived results have been directly transferred to the breeding sector, where these findings have been used for the designing of functional markers for the characterization and prediction of muscat flavor, berry color, and seedlessness.
Read all in: https://doi.org/10.1016/j.plantsci.2022.111539

Javier Tello graduated in Food Science and Technology in 2009 at the Universidad Autónoma de Madrid (Spain), where he also got his doctorate in Biology and Food Sciences in 2016. He started his research activities in grape and wine sciences at IMIDRA (Madrid, Spain), where he studied the genetic diversity of yeast populations in musts and wines from local wineries through molecular approaches. Then, he moved to the Institute of Grapevine and Wine Sciences (ICVV, La Rioja, Spain) to explore the genetic basis of grapevine bunch and berry traits through an integrated approach. It included the use of association mapping to detect significant associations between genetic variations and phenotypic diversity. After this period, he first moved to the Universität für Bodenkultur Wien (Tulln an der Donau, Austria) and then to the Amélioration Génétique et Adaptation des Plantes Méditerranéennes et Tropicales Research Institute (AGAP) of INRAE (Montpellier, France) for two consecutive post-doctoral stages. During these periods, he increased his skills in bioinformatics, genetic mapping approaches, statistics, and plant physiology. Likewise, he was involved in the designing of several high-throughput tools to ease phenotypic data acquisition. Since 2019, he is a post-doctoral researcher at the Grapevine Genetics and Genomics Research Group of the ICVV, where he is involved in different projects focused on the characterization of worldwide grapevine genetic resources, and on the understanding of the molecular basis of traits of agronomic relevance. He has participated in 12 research projects and in 15 contracts with national and international companies, contributing to their successful achievement by leading specific objectives. This work derived in the publication of 30 indexed scientific publications, two book chapters, and several technical articles for specialized journals.

Javier Ibáñez graduated in Biological Sciences in 1988, obtained a Bachelor’s Degree in 1990 and obtained a doctorate in Biological Sciences in 2000, all at the Complutense University of Madrid. He began working with the grapes at IMIDRA (Madrid) in 1995, in order to apply molecular analysis to the study of vine varieties. He pioneered in Spain DNA and statistical tools for varietal identification and characterization, and participated in the European networks that led this research, collaborating in the most significant works in this field. Since 2004 he also became interested in the genetic basis of natural variation for reproductive traits in grapevine, identifying the genes responsible for traits of interest in the berry and the bunch. In 2008 he combined his research activity at IMIDRA with teaching activity as Associate Professor in the Department of Genetics of the Complutense University of Madrid. In 2009 he joined the CSIC as Tenured Scientist at the Institute of Grapevine and Wine Sciences (ICVV, La Rioja), co-leading the Grapevine Genetics and Genomics research group (Vitigen) and deepening the study of the genetic diversity of the grapevine and its reproductive development. The latest projects that he has leaded have focused mainly on the characterization of the somatic variation of the vine, the genetics of the compactness of the bunch, and the genetic diversity in cultivated and sylvestris vines.In this time, he has led or participated in 46 research projects and 28 contracts with regional, national and international companies. He has supervised four doctoral theses and has 119 scientific publications (64 indexed), 13 books or book chapters and 22 popularization publications related to the exposed subject. He has been a guest speaker at 33 events, contributing with 84 scientific communications in international conferences and 54 in national ones.
- References:
Tello, J., Ibáñez, J. (2023). Review: Status and prospects of association mapping in grapevine. Plant Science 327, 111539. - Fournier-Level, A., Le Cunff, L., Gomez, C., Doligez, A., Ageorges, A., Roux, C., Bertrand, Y., Souquet, J.M., Cheyner, V., This, P. (2009). Quantitative genetic bases of anthocyanin variation in grape (Vitis vinifera L. ssp. sativa) berry: a quantitative trait locus to quantitative trait nucleotide integrated study, Genetics 183, 1127–1139.
- Emanuelli, F., Battilana, J., Costantini, L, Le Cunff, L. Boursiquot, J.M., This, P., Grando, M.S. (2010) A candidate gene association study on muscat flavor in grapevine (Vitis vinifera L.), BMC Plant Biology 10, 241.
- Royo, C., Torres-Pérez, R., Mauri, N., Diestro, N., Cabezas, J.A., Marchal, C., Lacombe, T., Ibáñez, J., Tornel, M., Carreño, J., Martínez-Zapater, J.M., Carbonell-Bejerano, P. (2018) The major origin of seedless grapes is associated with a missense mutation in the MADS-box gene VviAGL11, Plant Physiology 177, 1234–1253.

