By Anabella Varela, Federico Berli, Carlos Marfil
Genetic polymorphism is the main source of variability in a vineyard, were the producer usually grows different clones of a same cultivar. Clonal selection and vegetative propagation in viticulture determine low genetic variability among grapevine clones, although it is common to observe diverse phenotypes because of the broad phenotypic plasticity this crop presents. The term phenotypic plasticity refers to the range of phenotypes that a genotype expresses in different environments.
And what about epigenetics? The epigenetic polymorphism is another source of variability. These mechanisms include cytosine DNA methylation, histone post-translational modifications, and small RNAs regulation (Norouzitallab et al. 2019). In our work we will focus in the first of those mechanisms.
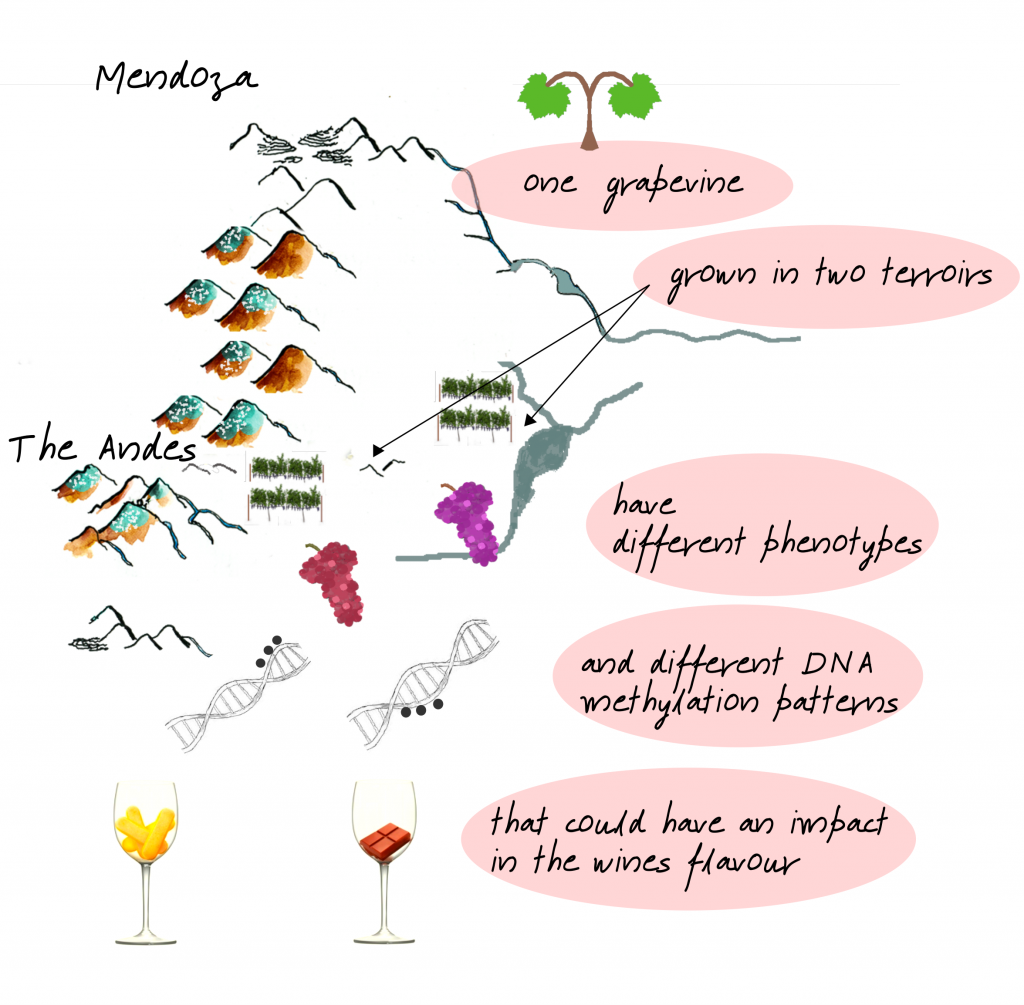
There is evidence that differences in DNA methylation play a role in phenotype traits in the model plant Arabidopsis thaliana (Johannes et al. 2009) and in the adaptation of perennial species with low genetic variation to changing environments; as in Pinus pinea L. (Sáez-Laguna et al. 2014) and Populus tremuloides (Ahn et al. 2017). Also, previous studies showed changes in grapevine DNA methylation induced by harsh environmental conditions like in vitro cultivation (Baránek et al. 2015; Schellenbaum et al. 2008), high ultraviolet-B and C radiation (UV-B and UV-C) (Marfil et al. 2019; Tyunin and Kiselev 2016), and kaolin treatments (Bernardo et al. 2017). Moreover, Xie et al. (2017) analyzed the molecular diversity of cv. Shiraz in 22 vineyards of Australia and observed low genetic differentiation, but a high level of epigenetic variation between vineyards. Similarly, we previously found that UV-B, water restriction and abscisic acid (ABA) treatments induced changes in DNA methylation patterns of cv. Malbec (Marfil et al. 2019). We showed that DNA methylation patterns were mitotically heritable and associated with the hydroxycinnamic acids accumulation in the early fruit shoots. All this evidence let us assume that environmental signals may induce epigenetic changes altering gene expression and phenotype. The concept of terroir, a popular French term frequently used in the wine industry, refers to the complex interactions between the genotype, the environment and human cultural practices. For a better understanding of the terroir effect on wine sensory attributes the inclusion of epigenomic data is necessary to know the mechanism of how the environment affects the grapevine phenotypes.
To put the study in a real context, we analyzed the effects on phenotypic traits and epigenome of three Vitis vinifera cv. Malbec clones (clone 01, 04 and 10) cultivated in two contrasting vineyards of Mendoza, Argentina: Agrelo and Gualtallary. Gualtallary vineyard was more humid and colder than Agrelo during the 2016/2017 season, in line with the historic records.
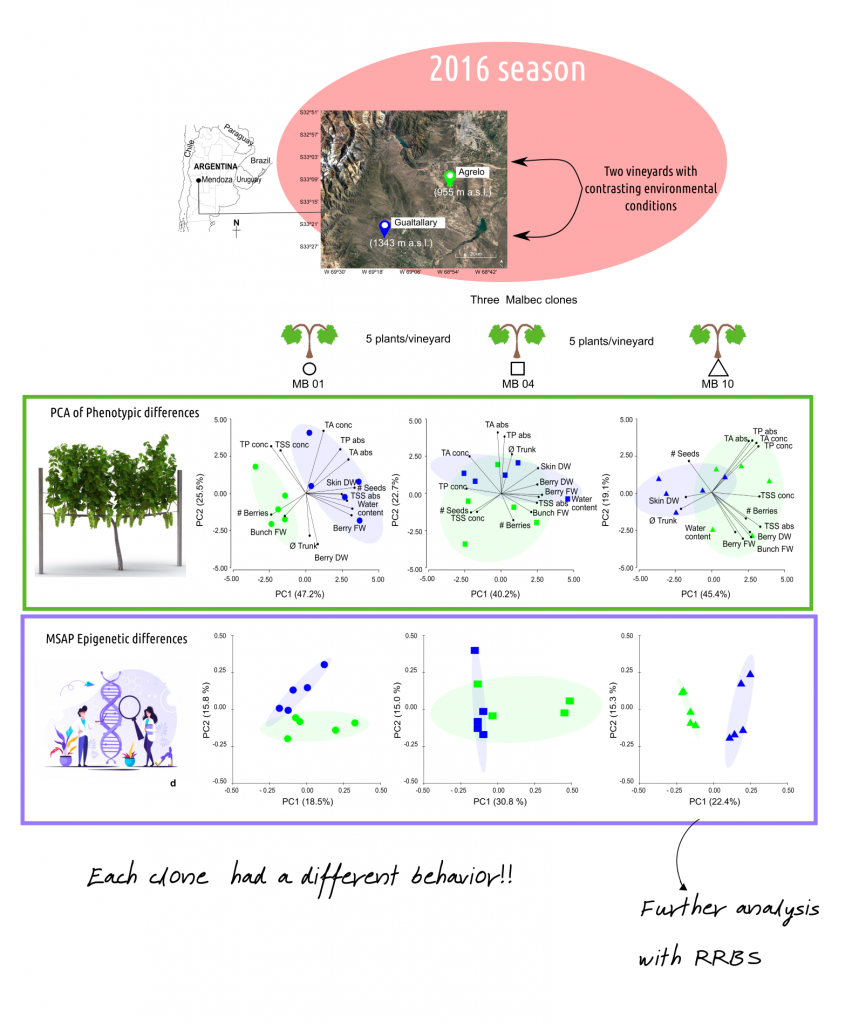
Sixteen phenotypic traits were measured: trunk diameter (at 65 cm from ground level), bunch fresh weight (FW), number of berries per bunch (# berries), berry FW, berry dry weight (DW; dried at 60 °C to a constant weight), berry water content (estimated as berry FW-berry DW), berry total soluble solids (TSS) in concentration (TSS conc) and TSS in a per berry basis (TSS abs), number of seeds per berry (# seeds), berry skin DW, berry skin total anthocyanins (TA conc and TA abs), berry skin polyphenolic content (TP conc and TP abs), specific leaf area (SLA), and leaf DW. Anonymous genome regions were analyzed using methylation-sensitive amplified polymorphism (MSAP) markers from aplical meritems DNA extractions. This enzymatic technique allows us to detect specific DNA methylation patterns trough anonymous sites from the genome and to compare the epigenetic polymorphism among plants. The clone that presented the clearer MSAP differentiation between vineyards was selected and analyzed through reduced representation bisulfite sequencing (RRBS). The RRBS is a high-throughput technique for analyzing the genome-wide methylation profile on a nucleotide level. It allows us to detect the identity of different methylated regions of the genome.
Our results showed phenotypic variability among clones and sites, where MB01 showed a clear clustering based on the vineyard with fruit yield traits associated with Agrelo, a partial differentiation between vineyards in MB10 with Skin DW associated with Gualtallary and no clustering according to vineyard for MB04. Also, the PCoA analysis of epigenetic polymorphism showed different methylation patterns associated with the vineyard for MB01 and MB10, being more notorious in MB10, while plants of MB04 clone showed no differences between vineyards. Correlation between MSAP epigenetic and phenotypic variability was detected. A small but significant linear correlation was observed (R= 0.15, p value: 0.002) when the entire sets of data for the three clones were considered. When clones were analyzed separately, only MB10 showed a linear correlation between epigenetic and phenotypic variability.
RRBS analysis of MB10 also clustered plants by vineyards and significant correlations were observed for RRBS methylome dataset and Skin DW/TSS abs and Skin DW/TSS abs/TSS conc subgroups. Twenty-nine differentially methylated regions between vineyards were identified and associated to genes and/or promoters and some differentially methylated genes could have some relationship with the phenotypic differences observed.
Which are the main conclusions from our work?
First that in Malbec the phenotypic plasticity and epigenetic modulation is clone dependent. The environment had a different effect in the phenotype of the clones despite their low genetic inter-clone variability and also had a different effect in the methylation patterns observed. Thus, DNA methylation pattern variations may be inter-related to DNA sequence variation (Li et al. 2008).
Secondly, MSAP and RRBS results reinforced the idea that the environment shapes the epigenome somehow. These results are consistent with previews research were MSAP methylation patterns were region dependent (Xie et al. 2017)
Also, that the low MSAP epigenetic and phenotypic correlation could probably be due to many small epigenetic changes across the genome rather than few changes with a radical impact. And that DNA methylation is probably not associated with the phenotypic plasticity in general, but rather with a subset of traits.
For more detailed information read https://link.springer.com/article/10.1007/s00299-020-02617-w

Anabella Varela 
Federico Berli 
Carlos Marfil 
Anabella Varela. Masters degree in Vititulture and Oenology in the University of Pavoda, Udine and Verona. PhD student working on Vitis vinifera epigenetics in the Instituto de Biología Agrícola de Mendoza, CONICET-Universidad Nacional de Cuyo, Argentina. Advisors: Carlos Marfil and Federico Berli.
Federico Berli. Professor of Plant Biochemistry and Researcher in the Instituto de Biología Agrícola de Mendoza, CONICET-Universidad Nacional de Cuyo, Argentina. His research is focused on the effects of environmental factors on the grapevine physiology. He is author of 21 papers in international scientific journals. Mail: fberli@fca.uncu.edu.ar
Carlos Marfil. Professor of Genetics and Researcher in the Instituto de Biología Agrícola de Mendoza, CONICET-Universidad Nacional de Cuyo, Argentina. We develop researches related to the study of epigenetic variability as a source of phenotypic novelties in plants: we are interested in describing factors that generate epigenetic variability and the stability and importance of this variability in processes of acclimatization, adaptation and plant diversification. To study the environmental induced epigenetic variation we use as experimental models germplasm from agro-ecosystems (vine) and from natural ecosystems (wild potato species).
References
- Ahn, J., Franklin, S. B., & Douhovnikoff, V. (2017). Epigenetic variation in clonal stands of aspen. Folia Geobotanica, (January 2016). https://doi.org/https://doi.org/10.1007/s12224-017-9308-x
- Baránek, M., Čechová, J., Raddová, J., Holleinová, V., Ondrušíková, E., & Pidra, M. (2015). Dynamics and reversibility of the DNA methylation landscape of grapevine plants (Vitis vinifera) stressed by in vitro cultivation and thermotherapy. PLoS ONE, 10(5), e0126638. https://doi.org/10.1371/journal.pone.0126638
- Bernardo, S., Dinis, L.-T., Luzio, A., Pinto, G., Meijón, M., Valledor, L., … Moutinho-Pereira, J. (2017). Kaolin particle film application lowers oxidative damage and DNA methylation on grapevine ( Vitis vinifera L.). Environmental and Experimental Botany, 139, 39–47. https://doi.org/10.1016/j.envexpbot.2017.04.002
- Johannes, F., Porcher, E., Teixeira, F. K., Saliba-Colombani, V., Simon, M., Agier, N., … Colot, V. (2009). Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genetics, 5(6), e1000530. https://doi.org/10.1371/journal.pgen.1000530
- Li, Y., Shan, X., Liu, X., Hu, L., Guo, W., & Liu, B. (2008). Utility of the methylation-sensitive amplified polymorphism ( MSAP ) marker for detection of DNA methylation polymorphism and epigenetic population structure in a wild barley species (Hordeum brevisubulatum). The Ecological Society of Japan, 23, 927–930. https://doi.org/10.1007/s11284-007-0459-8
- Marfil, C., Ibañez, V., Alonso, R., Varela, A., Bottini, R., Masuelli, R., … Berli, F. (2019). Changes in grapevine DNA methylation and polyphenols content induced by solar ultraviolet-B radiation , water de fi cit and abscisic acid spray treatments. Plant Physiology and Biochemistry, 135, 287–294. https://doi.org/10.1016/j.plaphy.2018.12.021
- Norouzitallab, P., Baruah, K., Vanrompay, D., & Bossier, P. (2019). Can epigenetics translate environmental cues into phenotypes ? Science of the Total Environment, 647, 1281–1293. https://doi.org/10.1016/j.scitotenv.2018.08.063
- Sáez-Laguna, E., Guevara, M.-Á., Díaz, L., Sánchez-Gómez, D., Collada, C., Aranda, I., & Cervera, M.-T. (2014). Epigenetic Variability in the Genetically Uniform Forest Tree Species Pinus pinea L. PLoS ONE, 9(8), 1–10. https://doi.org/10.1371/journal.pone.0103145
- Schellenbaum, P., Mohler, V., Wenzel, G., & Walter, B. (2008). Variation in DNA methylation patterns of grapevine somaclones (Vitis vinifera L.). BMC Plant Biology, 8(1), 78. https://doi.org/10.1186/1471-2229-8-78
- Tyunin, A. P., & Kiselev, K. V. (2016). Alternations in VaSTS gene cytosine methylation and t -resveratrol production in response to UV-C irradiation in Vitis amurensis. Plant Cell, Tissue and Organ Culture (PCTOC), 124, 33–45. https://doi.org/10.1007/s11240-015-0872-6
- Xie, H., Konate, M., Sai, N., Tesfamicael, K. G., Cavagnaro, T., Gilliham, M., … Lopez, C. M. R. (2017). Global DNA Methylation Patterns Can Play a Role in Defining Terroir in Grapevine (Vitis vinifera cv. Shiraz). Frontiers in Plant Science, 8, 1860. https://doi.org/10.3389/fpls.2017.01860

