By Mark Whitty
Background
Have you ever wondered just what causes the structure of grape bunches to vary? The Smart Robotic Viticulture group at UNSW Australia has been developing methods for measuring and modelling 3D grape bunches based on image analysis. This enables large scale phenotyping which in turn lays the foundation for studies into the relationships between bunch architecture and wine quality or disease potential.
What we did
Initial work in this space by Dr Scarlett Liu was to use the predominantly the outline of a bunch to constrain the arbitrary placement of exterior berries (Liu, 2015). We then extended it to include visible berries detected in the centre of the bunch and improved its’ robustness to bunch colour (Liu, 2020a) as well as turn the algorithm into an app (Liu, 2020b). However, while these initial approaches use actual bunch shapes, they have no appreciation of the actual rachis structure which connects the berries.
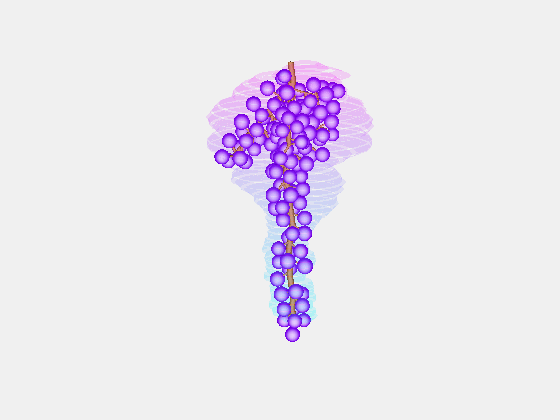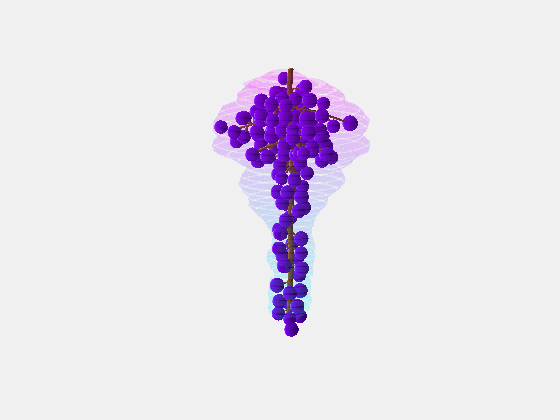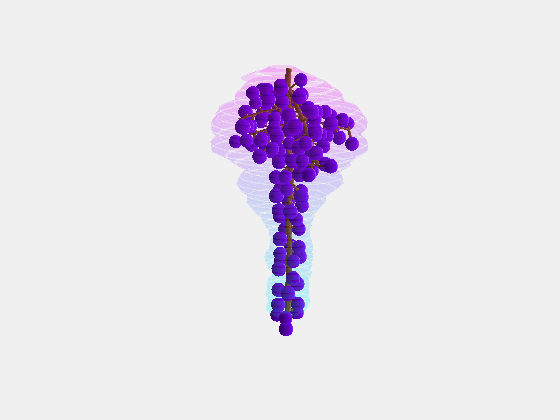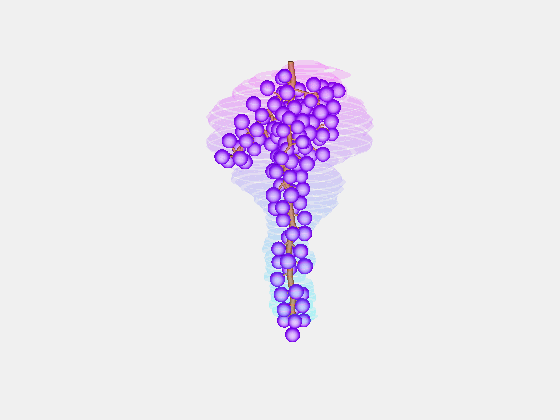
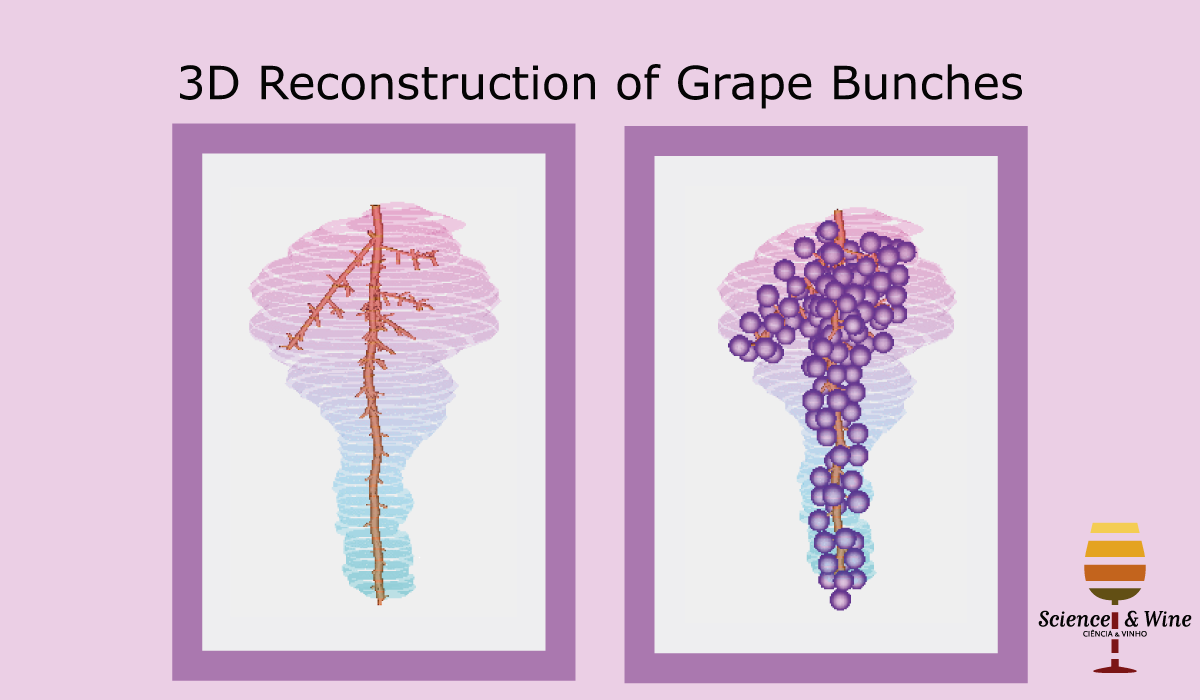
Bolai Xin then began by measuring and documenting the full rachis structure of a few dozen Pinot Noir and Merlot bunches – with the kind assistance of Damian Martin and his team in Blenheim at The New Zealand Institute for Plant and Food Research and Justin Jarrett from See Saw Wines.
From this we constructed a stochastic model of each cultivar, with the existence and length of every internode represented probabilistically, something which has not been done previously.
Our key contribution (Xin, 2020) has been to combine this with an actual image of a bunch, using the outline to form a convex hull which restricts the model as it reconstructs the full architecture stochastically. This enables the bunch architecture to be true to both the stochastic model for that variety as well as have a unique shaped based on the image.
Thus, given a single image of a bunch, we can generate a representative estimate of its 3D structure including all internodes.
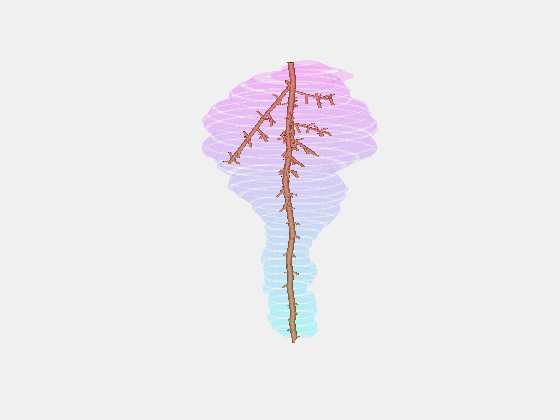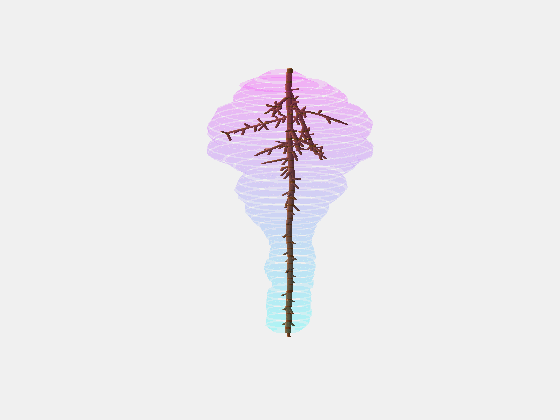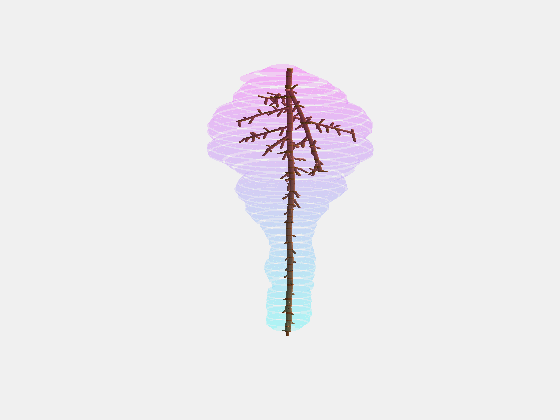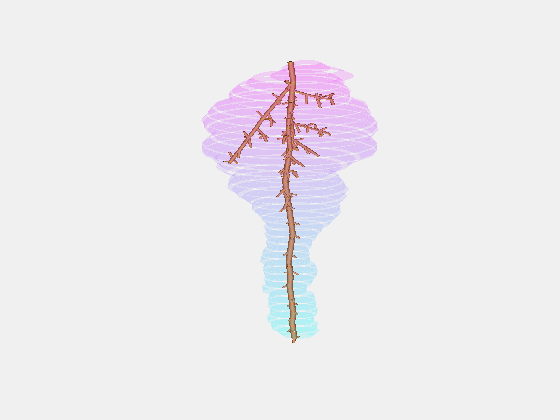
What we are doing now
We have since developed a method which not only constrains the stochastic model by its outline but links it explicitly to all visible berries. This then enables the invisible portion of the bunch to be reconstructed in a manner unique to that cultivar and that image. Stay tuned for details!
What comes next
Once reconstructed, the 3D bunch model architecture can be used to gain insight into bunch variation.
Want to know more about the architecture of your grape bunches? Contact us!

Dr Mark Whitty is a senior lecturer in the School of Mechanical and Manufacturing Engineering at the University of New South Wales (UNSW Sydney), Australia. He leads the Smart Robotic Viticulture team at UNSW and his research interests include digital viticulture, digital agriculture, agricultural robotics, and 3D point cloud processing. He has been successful with both nationally competitive grants and engagement with industry internationally in the agricultural sector. He received a Ph.D. in Mechatronic Engineering from UNSW. He is a member of the IEEE. Contact him at m.whitty@unsw.edu.au.
References
- Liu S; Whitty M; Cossell S, 2015, ‘A Lightweight Method for Grape Berry Counting based on Automated 3D Bunch Reconstruction from a Single Image’, in A Lightweight Method for Grape Berry Counting based on Automated 3D Bunch Reconstruction from a Single Image, ICRA, International Conference on Robotics and Automation (IEEE), Workshop on Robotics in Agriculture, Seattle, 30 May 2015 – 30 May 2015.
- Liu S; Zeng X; Whitty M, 2020a, ‘A vision-based robust grape berry counting algorithm for fast calibration-free bunch weight estimation in the field’, Computers and Electronics in Agriculture, vol. 173, http://dx.doi.org/10.1016/j.compag.2020.105360Liu S; Zeng X; Whitty M, 2020b, ‘3DBunch: A novel iOS-smartphone application to evaluate the number of grape berries per bunch using image analysis techniques’, IEEE Access, vol. 8, pp. 114663 – 114674, http://dx.doi.org/10.1109/access.2020.3003415
- Xin B; Liu S; Whitty M, 2020, ‘Three-dimensional reconstruction of Vitis vinifera (L.) cvs Pinot Noir and Merlot grape bunch frameworks using a restricted reconstruction grammar based on the stochastic L-system’, Australian Journal of Grape and Wine Research, vol. 26, pp. 207 – 219, http://dx.doi.org/10.1111/ajgw.12444

[…] Science & Wine: 3D reconstruction of grape bunches […]