By Warren Albertin, Patricia Ballestra, Marguerite Dols-Lafargue, Isabelle Masneuf-Pomarede
Brettanomyces bruxellensis is considered as THE wine spoilage yeast. ‘Brett’ ruins wines all around the world, producing volatile phenols whose aromas are described as leather, horse sweat or stable and could mask the fruity perception of the red wines. Table wines as well as ‘grand crus’ can be spoiled, and the current treatments (sulfur dioxide, chitosane treatment, filtering, etc.) have mitigated effects since up to 25% of red wines are contaminated (Gerbaux et al. 2000, Bulletin de l’OIV). An important part of the scientific and technical literature in the field of wine microbiology is dedicated to ‘Brett’, but sometimes with contradictory results and inconsistent recommendations. Surprisingly, the wine foe is a brewer’s friend: kombucha and some beers are brewed using Brett strains, and its use for bioethanol production is explored (Steensels et al. 2015, Int J Food Microbiol).
These past years, the extraordinary diversity of Brett species was unveiled, first through the whole genome sequencing of a few strains (Crauwels et al. 2014, Applied and Environmental Microbiology; Curtin et al. 2012, PLoS One; Fournier et al. 2017, G3 (Bethesda); Gounot et al. 2019, bioRxiv; Olsen et al. 2015, GigaScience), then via the genetic characterization of hundreds of Brett from >20 countries and various substrates. These works showed the existence of at least 6 sub-populations, some of them encompassing diploid individuals (two sets of chromosomes) and other triploid individuals (three sets of chromosomes). In addition, these groups are strongly associated with processes, with beer, kombucha, bioethanol and tequila strains clustering in specific sub-populations (Avramova et al. 2018, Scientific reports).
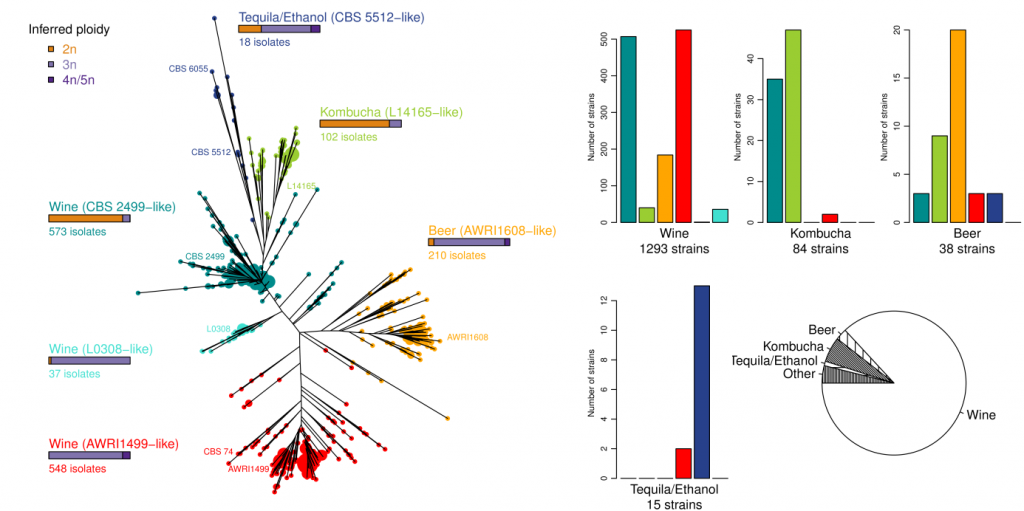
Isolates from wines or wine-environment are found in five out of the six subpopulations (Cibrario et al. 2019, Plos One). Brett distribution varies greatly depending on the vineyard: two main groups are found in Bordeaux (Wine 2N and 1st Wine 3N), while Portugal shows a perfect distribution among the five subpopulations for example.
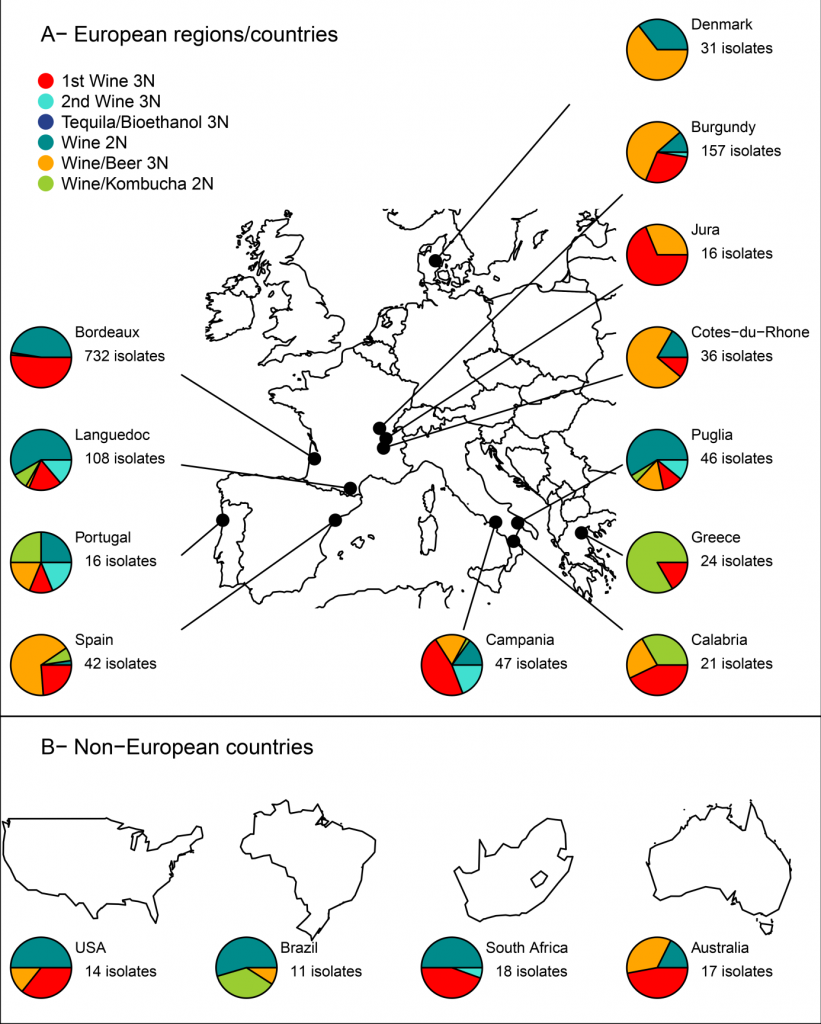
While variety is the spice of life, when it comes to Brett, diversity may be the ban of wine. The different sub-populations have distinct characteristics: red and turquoise groups were described as tolerant/resistant to sulfite treatments (Avramova et al. 2018, Frontiers in Microbiology; Dimopoulou et al. 2019, Food Microbiol), and preliminary results suggest variation in bioadhesion abilities that may impact the efficiency of cleaning/disinfection procedures (Dimopoulou et al. 2019, Ann Microbiol). Indeed, the analysis of wine isolates from very old vintages (as old as 1909) revealed that some genetically identical strains could persist for decades within the same winery. Furthermore, identical strains are found across continents, perhaps as a result of international shipping of wine and/or equipment. Finally, the proportions of the different populations have shifted globally over the last decades, perhaps as a result of evolving winemaking practices, such as increased use of sulfur dioxide as a preservative, or a trend toward higher alcohol content (Cibrario et al. 2019, Plos One). By contrast, volatile phenol production is ubiquitous within the species, as well as the ability to use various sugars to support growth (Cibrario et al. in press, Food Microbiology). However, not all the wines display the same ability to promote Brett growth (Cibrario et al, 2019, Am. J. Enol. Vitic.).
Future work on Brett will have to take into account this incredible diversity to better control wine spoilage.
Acknowledgements: These results have been achieved thanks to the collaboration of various international laboratories/institute: the universities of Bordeaux, Strasbourg, Brest, Burgundy in France, Foggia and Naples, Tarragona, Adelaide, Thessaloniki, the AWRI (Australian Wine Research Institute). Several organizations also provided Brett isolates: ICV, France; Inter Rhône, France; Technical University of Lisbon, Portugal; Universidade Federal de Pernambuco, Brazil; IFV, France; Microflora, France; University of Western Ontario, Canada; University of Food Technology, Bulgaria.

Warren Albertin is Assistant Professor in the Oenology research unit, Institute of Vine & Wine Science (ISVV), University of Bordeaux. Her research activities focus on the study of non-conventional wine yeasts, such as Brettanomyces bruxellensis, Torulaspora delbrueckii or Lachancea thermotolerans using large-scale population genetics and phenotyping approaches.

Patricia Ballestra is Assistant Professor in the Oenology research unit, Institute of Vine & Wine Science (ISVV), University of Bordeaux. She contributes to the characterization of wine spoilage microorganisms, focusing on Brettanomyces bruxellensis yeasts and Oenococcus oeni bacteria.

Isabelle Masneuf-Pomarede is professor of enology and she is at the head of the scientific department of Bordeaux Sciences Agro. Isabelle Masneuf-Pomarede conducts research on wine microbiology, especially of wine yeasts including fermentative and spoilage yeast.

Marguerite Dols-Lafargue is professor of biochemistry in Bordeaux INP and researcher in the Oenology research unit, Institute of Vine & Wine Science (ISVV), University of Bordeaux. Her main research topic deals with wine microbe (yeasts such as Brett or bacteria) carbohydrate metabolism (catabolism and polysaccharide production).
Bibliography
- Avramova M, Cibrario A, Peltier E, Coton M, Coton E, Schacherer J, Spano G, Capozzi V, Blaiotta G, Salin F, Dols-Lafargue M, Grbin P, Curtin C, Albertin W, Masneuf-Pomarede I (2018) Brettanomyces bruxellensis population survey reveals a diploid-triploid complex structured according to substrate of isolation and geographical distribution. Scientific reports 8 doi:10.1038/s41598-018-22580-7
- Avramova M, Vallet-Courbin A, Maupeu J, Masneuf-Pomarède I, Albertin W (2018) Molecular Diagnosis of Brettanomyces bruxellensis’ Sulfur Dioxide Sensitivity Through Genotype Specific Method. Frontiers in Microbiology 9 doi:10.3389/fmicb.2018.01260
- Cibrario A, Avramova M, Dimopoulou M, Magani M, Miot-Sertier C, Mas A, Portillo MC, Ballestra P, Albertin W, Masneuf-Pomarede I, Dols-Lafargue M (2019) Brettanomyces bruxellensis wine isolates show high geographical dispersal and long persistence in cellars Plos One doi:10.1101/763441
- Cibrario A, Miot-Sertier C, Paulin M, Bullier B, Riquier L, Perello MC, De Revel G, Albertin W, Masneuf-Pomarede I, Ballestra P, Dols-Lafargue M (in press) Brettanomyces bruxellensis phenotypic diversity, tolerance to wine stress and spoilage ability. Food Microbiology.
- Cibrario A., P. Ballestra, L. Riquier, G. de Revel, I. Masneuf-Pomarède, M. Dols-Lafargue. 2019 (in press) Cellar temperature strongly affects Brettanomyces bruxellensis population and volatile phenols production in Bordeaux aging wines. Am J enol vitic. doi: 10.5344/ajev.2019.19029
- Crauwels S, Zhu B, Steensels J, Busschaert P, De Samblanx G, Marchal K, Willems KA, Verstrepen KJ, Lievens B (2014) Assessing Genetic Diversity among Brettanomyces Yeasts by DNA Fingerprinting and Whole-Genome Sequencing. Applied and Environmental Microbiology 80:4398-4413 doi:10.1128/AEM.00601-14
- Curtin CD, Borneman AR, Chambers PJ, Pretorius IS (2012) De-Novo Assembly and Analysis of the Heterozygous Triploid Genome of the Wine Spoilage Yeast Dekkera bruxellensis AWRI1499. PLoS One 7 doi:10.1371/journal.pone.0033840
- Dimopoulou M, Hatzikamari M, Masneuf-Pomarede I, Albertin W (2019) Sulfur dioxide response of Brettanomyces bruxellensis strains isolated from Greek wine. Food Microbiol 78:155:163 doi:10.1016/j.fm.2018.10.013
- Dimopoulou M, Renault M, Dols-Lafargue M, Albertin W, Herry JM, Bellon-Fontaine MN, Masneuf-Pomarede I (2019) Microbiological, biochemical, physicochemical surface properties and biofilm forming ability of Brettanomyces bruxellensis. Ann Microbiol In press doi:10.1101/579144v1
- Fournier T, Gounot JS, Freel K, Cruaud C, Lemainque A, Aury JM, Wincker P, Schacherer J, Friedrich A (2017) High-Quality de Novo Genome Assembly of the Dekkera bruxellensis Yeast Using Nanopore MinION Sequencing. G3 (Bethesda) 7:3243-3250 doi:10.1534/g3.117.300128
- Gerbaux V, Jeudy S, Monamy C (2000) Étude des phénols volatils dans les vins de Pinot noir en Bourgogne vol 73.
- Gounot J-S, Neuvéglise C, Freel KC, Devillers H, Piškur J, Friedrich A, Schacherer J (2019) High complexity and degree of genetic variation in Brettanomyces bruxellensis population. bioRxiv:826990 doi:10.1101/826990
- Olsen RA, Bunikis I, Tiukova I, Holmberg K, Lotstedt B, Pettersson OV, Passoth V, Kaller M, Vezzi F (2015) De novo assembly of Dekkera bruxellensis: a multi technology approach using short and long-read sequencing and optical mapping. GigaScience 4:56 doi:10.1186/s13742-015-0094-1
- Steensels J, Daenen L, Malcorps P, Derdelinckx G, Verachtert H, Verstrepen KJ (2015) Brettanomyces yeasts — From spoilage organisms to valuable contributors to industrial fermentations. Int J Food Microbiol 206:24-38 doi:http://dx.doi.org/10.1016/j.ijfoodmicro.2015.04.005
@Post image adatpted from https://doi.org/10.3390/fermentation2030017