By Vera Quecini, Iraci Sinski, Patrícia S. Ritschel, Thor V. M. Fajardo
Resistance to pathogens is an important goal in the development of novel grapevine cultivars throughout the world by conventional and biotechnology-assisted breeding programs. Fungal and viral diseases cause direct losses in berry production, but also affect the quality of the final products. Methods for chemical control are available for fungus, but they increase the production costs, decrease fruit quality, and negatively affect culture sustainability. For viral pathogens, the situation is also complex, since it has to rely on the control of vector insects, if that is the way of transmission, or, on the replacement of highly colonized plants by new, clean ones. Thus, the introduction of genetic resistance is economically and environmentally desirable. Resistance to fungi and viruses is often found in wild or non-commercial Vitis genotypes, which simultaneously carry many undesirable traits to the progeny when used in crosses with elite cultivars. Recently, precision breeding strategies, such as genetic engineering and genome editing, allow the precise introduction of resistance characters in elite cultivars (Figure 1). However, the overall performance of the plants submitted to precision breeding techniques is not always as expected.
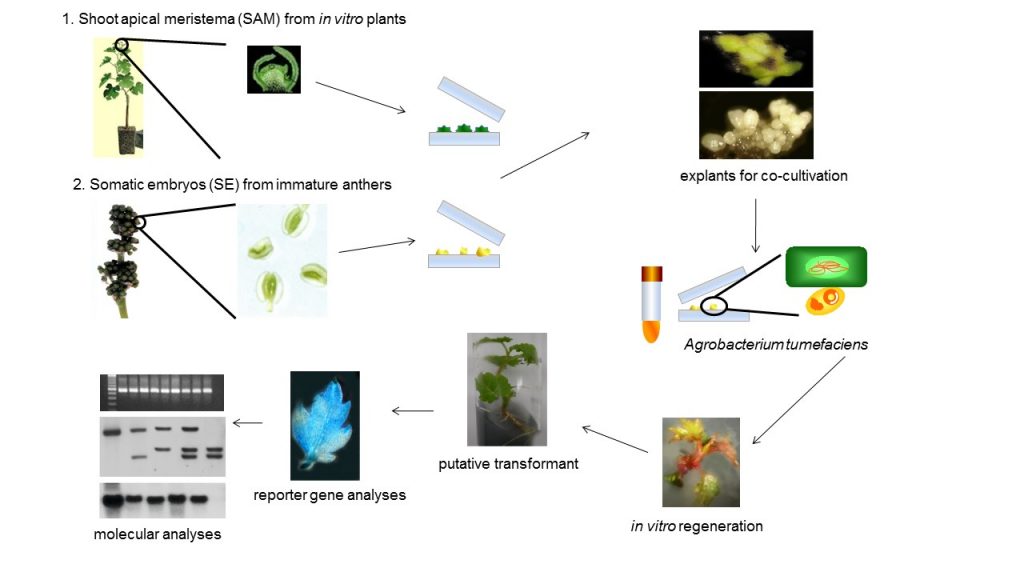
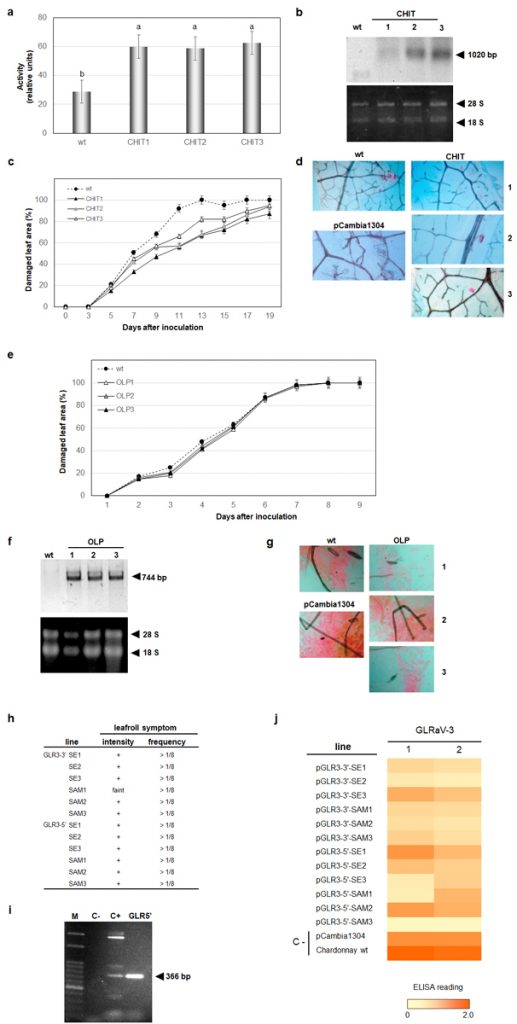
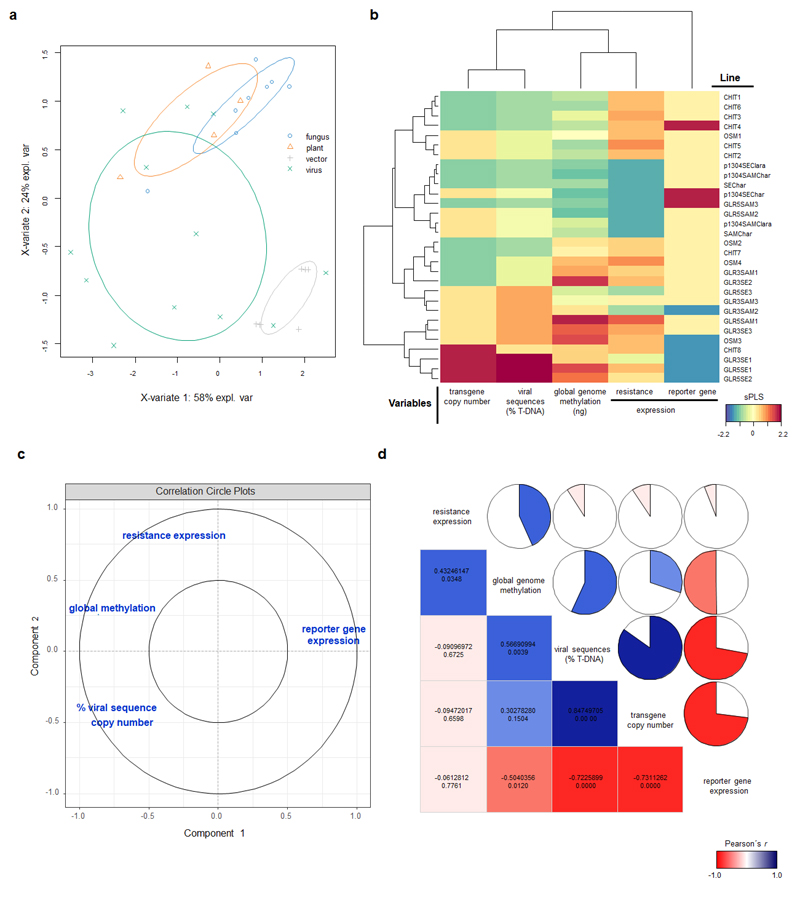
In the current work, we have generated genetically engineered grapevines that express proteins with antimicrobial properties against fungal pathogens or a virus-derived sequence, in a hairpin orientation, which triggers the specific degradation of an RNA essential to the formation of novel infective viral particles. As expected, the genetically engineered grapevines exhibited increased resistance to the pathogens, in comparison to the non-engineered plants, although the infection was still detected in the modified plants (Figure 2). Further investigation demonstrated that the plants carrying larger portions of sequences from viral origin exhibited higher levels of a structural genome modification called methylation, and lower levels of resistance (Figure 3). Methylation consists in the addition of methyl radicals to certain bases of the DNA and is considered an epigenetic modification, since it interferes with the activity of the DNA without modifying the sequence. Taken together, our results demonstrated that the introduction of sequences of viral origins into grapevine genome is associated to genome structural changes and reduced expression of resistance against pathogens, thus indicating that the effectiveness of resistance strategies employing sequences of viral origin is subject to epigenetic regulation in grapevine.
Those interested in a longer length report can download the working paper at:
https://link.springer.com/article/10.1007/s11248-018-0082-1
Vera Quecini
Graduated in Agricultural Engineering at the ESALQ, in the University of São Paulo, Brazil, where she also got her MSc. in Agricultural Microbiology and Doctorate in Genetics and Plant Breeding. Subsequently, she worked as post-doctoral researcher at the University of Wageningen, in the Netherlands, and at the University of Columbia, Missouri, in the United States. Currently, she is a researcher at the Grapevine and Wine Research Center of the Brazilian Agricultural Research Corporation (Embrapa).

Viraci Sinski
Graduated in Biology at the University of Caxias do Sul, Brazil.Currently, she is the responsible technician for the Tissue Culture and Biotechnology Laboratory at the Grapevine and Wine Research Center of the Brazilian Agricultural Research Corporation (Embrapa).

Patrícia S. Ritschel
Graduated in Agronomy at the University of Brasília and proceeded to get her MSc. in Genetics and Plant Breeding at Viçosa Federal University and Doctorate in Biological Sciences (Molecular Biology) at the University of Brasília. Currently, she is a researcher at the Grapevine and Wine Research Center of the Brazilian Agricultural Research Corporation (Embrapa). She is the head scientist for the grapevine breeding program at Embrapa and is responsible for the development of the novel cultivars of seedless (BRS Vitória, BRS Isis and BRS Melodia) and seeded (BRS Núbia) table grapes, juice- (BRS Carmem and BRS Magna) and wine-production (BRS Bibiana) varieties.

Thor V. M. Fajardo
Graduated in Agronomy at Viçosa Federal University, where he proceeded to get his MSc. in Phytopathology. Subsequently, he got his Doctorate at the University of Brasília and worked as post-doctoral researcher at CSIC-UPV, in Spain. Currently, he is a researcher at the Grapevine and Wine Research Center of the Brazilian Agricultural Research Corporation (Embrapa), working with plant virology, mainly focusing the detection and molecular characterization of viruses from grapevine and temperate fruit crops.
Reference:
- Dal Bosco D. et al. 2018. Expression of disease resistance in genetically modified grapevines correlates with the contents of viral sequences in the T-DNA and global genome methylation. Transgenic Research, v. 27 (4): 379–396. https://doi.org/10.1007/s11248-018-0082-1

