By Marina Bely, Warren Albertin, Cécile Miot-Sertier, Isabelle Masneuf-Pomarede
The use of Active Dry Yeast (ADY) as a fermentation starter is widely recommended to secure the process and avoid sluggish or stuck fermentation. In an opposite direction, there is an increasing demand from wine producers to use indigenous S. cerevisiae strains from their own vineyards with the aim of limiting oenological inputs during the process and to try to increase specificities associated with the local “terroir”. Indeed, the selection of yeast strains from indigenous populations for industrial development has been the subject of different studies (Di Maro et al., 2007; Settanni et al., 2012; Tofalo et al., 2009; Tofalo et al., 2014; Garofalo et al., 2018a). However, in some cases, spontaneous fermentations could lead to irregular wine quality due to lack of alcoholic fermentation control, and resulted in potential alterations to the final products. Thus, there is a need for new developments and guidelines to improve technological processes for exploiting indigenous microorganisms.
Some wineries traditionally elaborate wines with « pied de cuve » (PdC) (Clavijo et al., 2011, Li et al., 2012; Moschetti et al., 2016). By promoting the growth of fermentative strains in a small volume, this method is used to trigger alcoholic fermentation, especially for the first vats filled at the beginning of the harvest period. The PdC is based on a small volume of fermented musts prepared a few days before harvest and used to inoculate the subsequent batches of grape juice. To avoid possible off-flavours, several grapes are harvested and processed separately in order to select the lot with the best fermentation kinetics and without sensory defects. The implementation of PdC can then proceed using must from fermenting tanks to inoculate the next batch. This process permits an already adapted yeast population to enter the fresh grape must. Till now, very few studies have reported results on PdC inoculation, except in some specific case (high sugar concentration fermentation), different PdC protocols were shown to limit the amount of acetic acid of the resulting wines (Bely et al., 2005).
Thus, the implementation of the PdC method is still empirical in wineries. The main objective of this study was to evaluate the impact of PdC on the microbial, chemical and sensory characteristics of the wine.
PdC Protocol
Two PdC were managed from Ugni blanc (UgB) and Sauvignon blanc (SB) grape varieties in duplicates. Undamaged grapes (15 kg) were manually harvested for each variety 4 days before harvest. The grapes were crushed in clean containers. After 2 days of alcoholic fermentation between 18 and 20°C, ammonium phosphate and thiamine were added to reach the level of 200 mg l-1 (Bely et al., 1990) and 0.60 mg.l-1 respectively. The PdC were then kept until around 30-50 % of the alcoholic fermentation was reached. Finally, the PdC were subjected to sensory and chemical analyses before their use. The PdC were transferred into four 225 liters barrels which were filled with Sauvignon gris must (Sugars 231 g l-1, pH 3.35, Total acidity 6.1, Yeast Assimilable Nitrogen 133 mg l-1, Turbidity 51 NTU). Yeast Assimilable Nitrogen level was adjusted to 200 mg l-1 (Bely et al., 1990). PdC inoculum volume ratio was 1/20 of Sauvignon gris must. Two additional barrels were prepared as controls: one was inoculated with the commercial strains Zymaflore X5 at 200 mg l-1 , and the other was spontaneously fermented. Microbial sampling was carried out on the Sauvignon gris must before alcoholic fermentation and on the 6 fermented barrels when they reached 75 % of the fermentation. For PdC just before inoculation, 30 randomly chosen colonies were collected. For barrels at 75 %, 10 colonies were collected for further genetic analysis.
Results
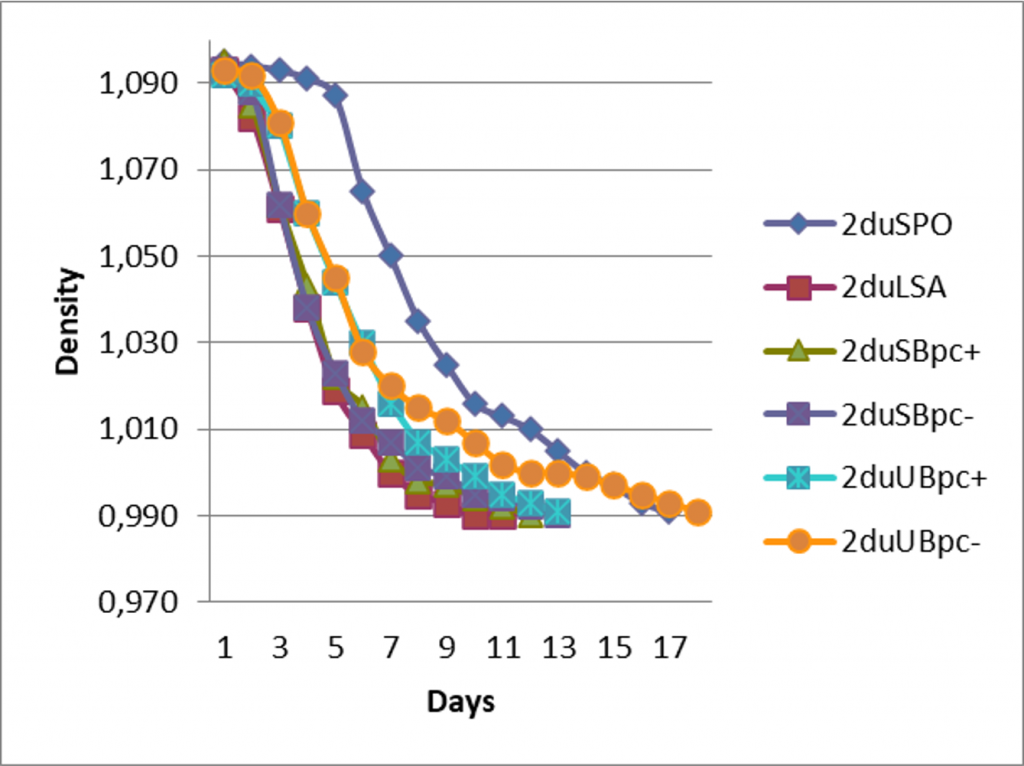
Just before the inoculation of the barrels, the different PdC modalities had similar population levels, from 2.107 to 6.107 UFC/mL for total yeast and 1.107 to 3.107 for non-Saccharomyces, thus suggesting that the non-Saccharomyces yeast population was the dominant one. The possibility of using the PdC to inoculate the barrels was further confirmed by sensory analysis, since no defect was detected. The fermentation kinetics were similar for the inoculated modality with X5 and for the PdC (Figure 1) except for one PdC UgB for which the fermentation was longer than for the spontaneous fermentation modality. Volatile acidity level (0.44 to 0.57 g l-1 acetic acid) was similar for all the modalities, except for PDC_UgB for which volatile acidity was high (0.61 g l-1 acetic acid), probably due to longer fermentation duration.
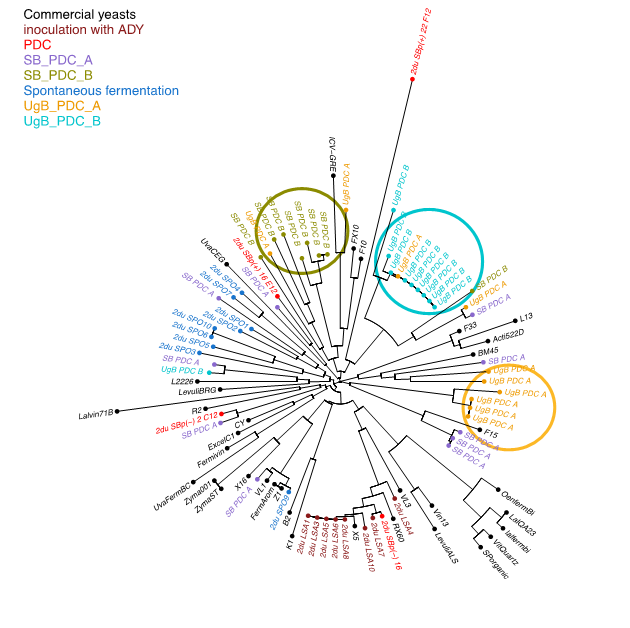
The microbiological analysis of the PdC revealed surprising results. First of all, the alcoholic fermentation of the PdC mainly occurred through non-Saccharomyces yeast; H.uvarum and S. cerevisiae were particularly rare. It was not possible to isolate S. cerevisiae strains in the 2 UgB PdC, probably due to the lack of ripening for the grapes when they were collected. Regarding the barrels, spontaneous fermentation showed high S. cerevisiae diversity, indicating that various strains grew leading to polyclonal populations during these fermentations, in contrast to ADY inoculation. For PdC inoculated barrels, clonal expansion of S. cerevisiae was highlighted during fermentation, especially for the UgB PdC, and to a lesser extent for the other modalities, with less than 50 % of the strains clustering together. The use of PdC seemed to enable the winemakers to select indigenous S. cerevisiae strains which differed from one modality to another, and to the S. cerevisiae population that occurred during spontaneous fermentation in the cellar. A dominant population, in line with what could be obtained by using ADY, may process alcoholic fermentation. The results seem to be uncertain since one modality failed in terms of fermentation kinetics and a high acetic acid level in the resulting wine. The preparation of different PdC modalities should thus lead to a more secure process.
Finally, a sensory analysis was performed and no significant differences were detected between all the modalities. Thus, in our experimental conditions, the use of PdC did not lead to distinct wines according to the origin of the yeast strains in the vineyard.

Isabelle MASNEUF-POMAREDE 
Warren ALBERTIN 
Cécile MIOT-SERTIER 
Marina BELY
Isabelle MASNEUF-POMAREDE is professor of enology at Bordeaux Sciences Agro and and in the Oenology research unit, Institute of Vine & Wine Science (ISVV), University of Bordeaux. She conducts research on wine microbiology, especially on wine yeasts community and its response to environmental changes and on the spoilage yeast Brettanomyces bruxellensis.
Warren ALBERTIN is Assistant Professor at Bordeaux INP and in the Oenology research unit, Institute of Vine & Wine Science (ISVV), University of Bordeaux. Her research activities focus on the study of non-conventional wine yeasts, such as Brettanomyces bruxellensis, Torulaspora delbrueckii or Lachancea thermotolerans using large-scale population genetics and phenotyping approaches.
Cécile MIOT-SERTIER is a technician at INRAE and in the Oenology research unit, Institute of Vine & Wine Science (ISVV), University of Bordeaux. Her main skills aremicrobiology, molecular biology, biochemistry, alcoholic and malolactic fermentations management.
Marina BELY is Assistant Professor in the Oenology research unit, Institute of Vine & Wine Science (ISVV), University of Bordeaux. Her research activities focus on wine yeast diversity, interactions between microorganisms, Bioprotection and fermentation management.
References
- Bely, M, I Masneuf, and D Dubourdieu. 2005. Influence of Physiological State of Inoculum on Volatile Acidity Production by Saccharomyces cerevisiae during High Sugar Fermentation.” Int J of Vine and Wine Sciences 39 (4).
- Clavijo A, Caldero ́n IL, Paneque P. Yeast assessment during alcoholic fermentation inoculated with a natural “pied de cuve” or a commercial yeast strain. 2011. World J Microbiol Biotechnol 7:1569–77.
- Di Maro E, Ercolini D, Coppola S. 2007. Yeast dynamics during spontaneous wine fermentation of the Catalanesca grape. Int J Food Microbiol. Jun 30;117(2):201-10.
- Garofalo C, Berbegal C, Grieco F, Tufariello M, Spano G, Capozzi V. 2018a. Selection of indigenous yeast strains for the production of sparkling wines from native Apulian grape varieties. Int J Food Microbiol. Nov 20;285:7-17.
- -Li E, Liu C, Liu Y. 2012. Evaluation of yeast diversity during wine fermentations with direct inoculation and pied de cuve method at an industrial scale. J Microbiol Biotechnol. Jul;22(7):960-6.
- Moschetti, G., O. Corona, R. Gaglio, M. Squadrito, A. Parrinello, L. Settanni, E. Barone, and N. Francesca. 2016. Use of Fortified Pied de Cuve as an Innovative Method to Start Spontaneous Alcoholic Fermentation for Red Winemaking. Australian J of Grape and Wine Research, 22, 36-45.
- Tofalo R, Perpetuini G, Fasoli G, Schirone M, Corsetti A, Suzzi G. 2014. Biodiversity study of wine yeasts belonging to the “terroir” of Montepulciano d’Abruzzo “Colline Teramane” revealed Saccharomyces cerevisiae strains exhibiting atypical and unique 5.8S-ITS restriction patterns. Food Microbiol. May 39: 7-12.
- Settanni, Luca, Ciro Sannino, Nicola Francesca, Rosa Guarcello, and Giancarlo Moschetti. 2012. Yeast Ecology of Vineyards within Marsala Wine Area (western Sicily) in Two Consecutive Vintages and Selection of Autochthonous Saccharomyces cerevisiae Strains. J of Bioscience and Bioengineering 114 (6): 606–14.

