By Giorgio Gambino and Paolo Boccacci
‘Nebbiolo’ (Vitis vinifera L.) is one of the most ancient and prestigious Italian grape cultivars. It is characterised by a great intra-varietal phenotypical polymorphism, resulting in 44 clonal selections officially registered in the Italian National Register of Grape Varieties (http://catalogoviti.politicheagricole.it/catalogo.php), which vary in morphological and physiological traits (e.g., leaf shape and size, shoot vigour, yield, phenolic content of juice at harvest). Its cultivation is widespread, although on limited surfaces, in different regions of the world (especially California and Australia), but the traditional cultivation area is limited to the hilly and mountainous zones of North-western Italy (Piedmont, Lombardy and Aosta Valley) where are produced 7 and 22 different wines certified with the DOCG (Denominazione di Origine Controllata e Garantita) and DOC (Denominazione di Origine Controllata) appellation, respectively.
The characteristics of the wines are strongly influenced by the must varietal composition, which directly impacts on the wine’s market price, especially in mono-varietal wines for which only one cultivar is used. Wine quality and value can be heavily modified if cultivars other than those allowed are employed. Therefore, the protection of local and regional wines with designation of origin labels is necessary for authenticity reasons, protecting consumers against frauds and speculations.
DNA typing has proved to be a valuable technique for accurately identifying cultivars due to its independence from external conditions and its high discriminating power. Among the available DNA markers, microsatellite or simple-sequence repeats (SSRs) are the markers of choice for grapevine fingerprinting. SSR markers have also been used to distinguish between cultivars using residual grape DNA extracted from mono- and multi-varietal musts and wines. Positive results were obtained in must analysis, but reproducibility problems were reported for the systematic authentication of experimental and commercial wines. The main limiting factors were the low DNA quality and quantity, mainly due to DNA degradation during the wine-making processes, reduction of DNA quantity by clarification and filtration of wines, presence of yeasts’ DNA, and PCR inhibitors, such as polyphenols, polysaccharides and proteins.
Single-nucleotide polymorphisms (SNPs) are considered the newest type of molecular marker for grapevine identification. SNPs can be employed to overcome the degradation limitations, allowing DNA amplification using more sensitive techniques, such as quantitative real-time polymerase chain reaction (qPCR). Although SNP polymorphism information content is lower compared with SSR, the high-throughput, next-generation sequencing technologies allow identifying a large number of SNPs in several genomes and develop panels of markers useful for cultivar identification, genetic diversity and mapping. In musts and wines, SNPs have recently been tested on experimental and commercial wines by qPCR, using specific TaqMan® probes or a high-resolution melting (HRM) approach.
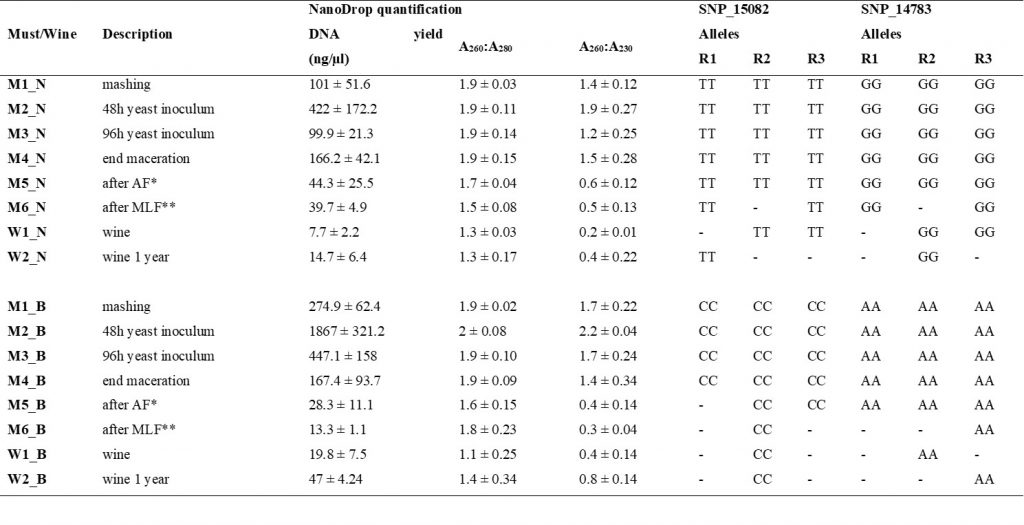
We developed and investigated the efficiency of SNP TaqMan® assays in the varietal authentication of ‘Nebbiolo’ musts and wines. Unlike SSRs, for which large databases are available, up to now there are still limited reference data for SNPs. However, using two set of data already published (Laucou et al., 2018; De Lorenzis et al., 2019) and analysing a group of genotypes ad hoc for the purposes of our work, using a Vitis18kSNP array containing 18,071 SNPs, we obtained a database of 1,157 different genotypes, from which ‘Nebbiolo’-specific SNPs were selected, and specific markers for other cultivars could be identified in the future. Only two markers (SNP_15082 and SNP_14783) were sufficient to distinguish ‘Nebbiolo’ from more than 1,100 genotypes. These markers were applied in the varietal identification of ‘Nebbiolo’ and ‘Barbera’ (used as an example of a non- ‘Nebbiolo’ cultivar) in musts and wines. In experimental vinifications, these SNPs TaqMan® assays correctly identified ‘Nebbiolo’ or ‘Barbera’ in all wine-making steps, including wines 1 year after bottling (Table 1). The high sensitivity of the assays allowed identifying, for the first time, mixtures of 1% of ‘Barbera’ in ‘Nebbiolo’ musts at the end of maceration (M4), blends of 10% in musts at the end of malolactic fermentation (M6) and contamination of 10–20% of ‘Barbera’ in ‘Nebbiolo’ wines (W1) (Figure 1). In commercial wines, the amplification efficiency of these SNPs was partially limited by the low amount of grapevine DNA and the presence of PCR inhibitors in DNA extracts. However, at least one SNP amplified correctly in all the wines tested. The TaqMan® genotyping protocol is a highly promising assay for varietal identification in wines for several reasons, including (i) high sensitivity and specificity in detecting DNA; (ii) reduced analysis time; and (iii) straightforward interpretation of results, even in non-specialised laboratories. The limited positive results obtained with commercial wines confirmed the difficulties reported in other works, and further improvements of the extraction techniques of nucleic acids from wine will be necessary.

Full-length article: Boccacci P., Chitarra W., Schneider A., Rolle L., Gambino G. (2020). Single-nucleotide polymorphism (SNP) genotyping assays for the varietal authentication of ‘Nebbiolo’ musts and wines. Food Chemistry 312, 126100. doi.org/10.1016/j.foodchem.2019.126100

Giorgio Gambino is a researcher at the Institute for Sustainable Plant Protection of the National Research Council of Italy (IPSP-CNR), Turin. He is mainly interested in the study of plant-pathogen and plant-environment interactions in grapevine and in the genetic markers for fingerprinting.

Paolo Boccacci is a research technician at the Institute for Sustainable Plant Protection of the National Research Council of Italy (IPSP-CNR), Turin. My research and technical support are mainly focused on plant breeding and plant-environment interactions.
References
- De Lorenzis, G., Chipashvili, R., Failla, O., & Maghradze, D. (2015). Study of genetic variability in Vitis vinifera L. germplasm by high-throughput Vitis18kSNP array: the case of Georgian genetic resources. BMC Plant Biology, 15, 154
- Laucou, V., Launay, A., Bacilieri, R., Lacombe, T., Adam-Blondon, A-F., Bérard, A., Chauveau, A., de Andrés, M. T., Hausmann, L., Ibáñez, J., Le Paslier, M-C., Maghradze, D., Martinez-Zapater, J. M., Maul, E., Ponnaiah, M., Töpfer, R., Péros, J-P., & Boursiquot, J-M. (2018). Extended diversity analysis of cultivated grapevine Vitis vinifera with 10K genome-wide SNPs. PLoS ONE, 13, e0192540


I hope that you continue to follow Science & Wine