By Renato L. Binati and Sandra Torriani
The variety of the microbiota naturally present in the vineyards is affected by several parameters, such as grape variety, location, climate, vintage, vineyard treatments, pruning techniques, growing stage and ripening (Varela & Borneman, 2017). In the spontaneous fermentation of wine, the process is carried out by a succession of that myriad of different species, together with the microbes colonizing the winery equipment. Yet, the inoculation of Saccharomyces cerevisiae starter cultures in grape musts became a common practice worldwide, due to its tolerance to the harsh conditions found in fermenting musts, excellent fermentation performance, and satisfactory aroma-related metabolism (Padilla et al., 2016). However, some critics of the extensive use of this approach claim that it could result in wines lacking complexity, typicity and distinction, which could presumably be augmented with native non-Saccharomyces yeasts as co-starters, due to their relevant enzymatic activities and production of metabolites of oenological significance (Jolly et al., 2006).
This study, graphically represented in Figure 1, used culture-dependent methods to set up a vast culture collection gathered from a wide range of Italian viti-vinicultural regions and assess the non-conventional yeast diversity, focusing on oenologically interesting species. The screening integrated genomic and technological features aiming to exploit the huge untapped potential of native microorganisms.
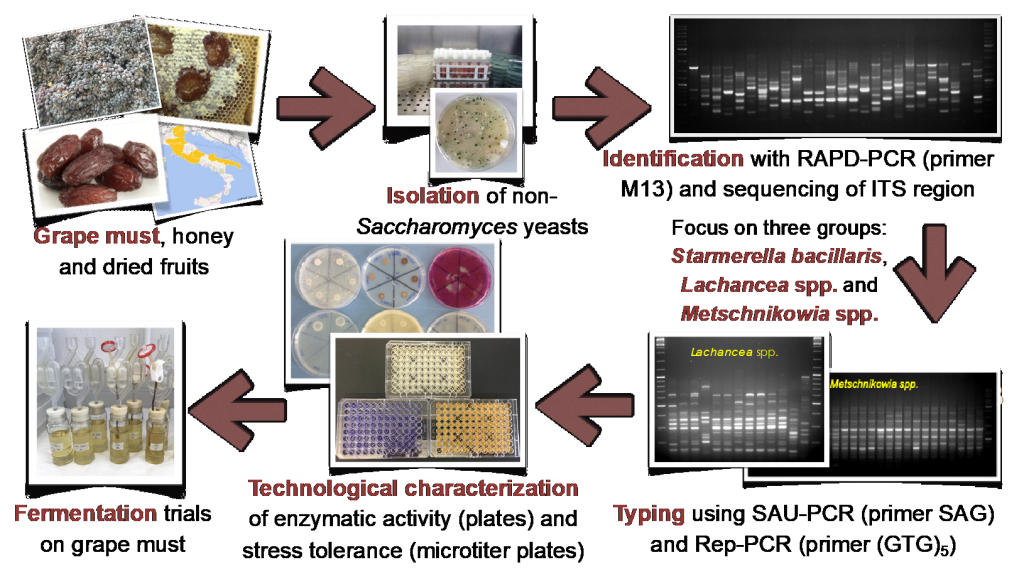
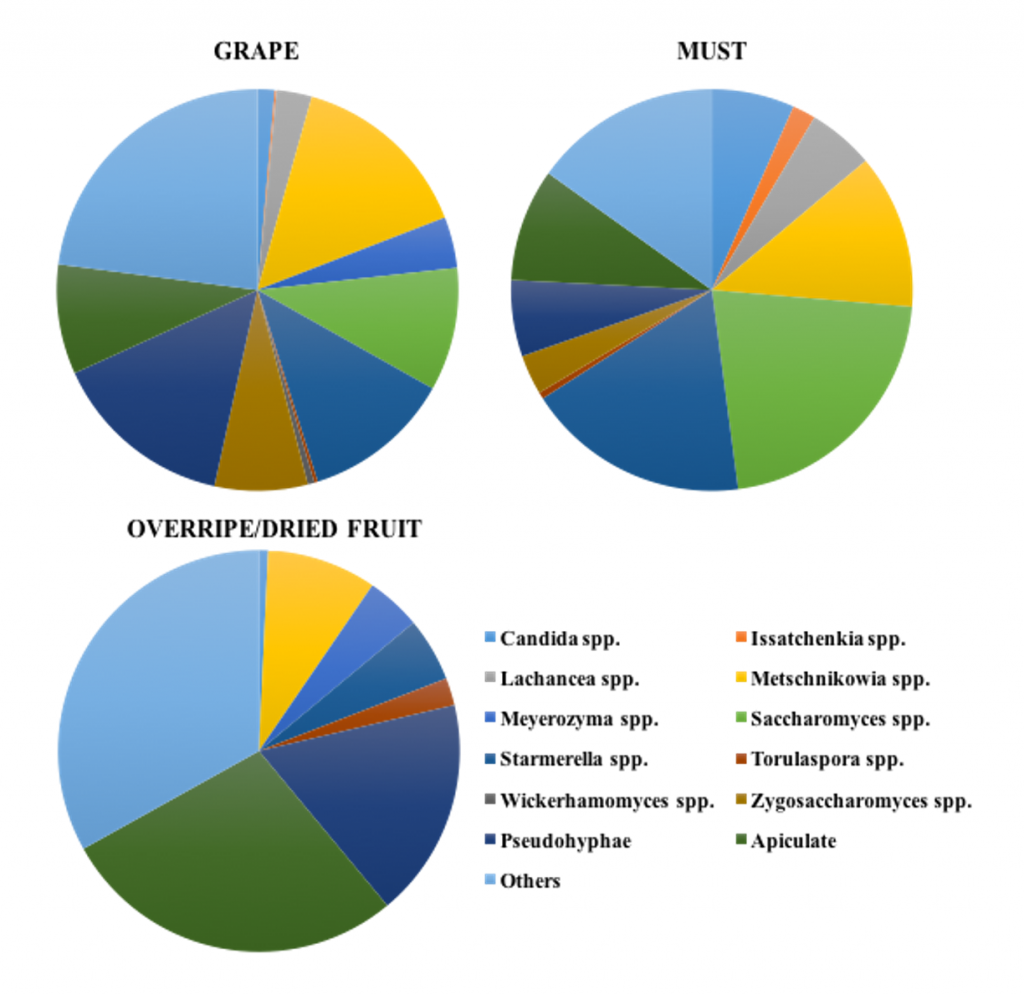
A high diversity of non-Saccharomyces yeasts was found after mining more than 400 isolates in 167 samples of grapes, grape musts and other high-sugar matrices, such as honey, overripe and dried fruits. Careful observation of yeast colony and cell morphology, combined with molecular methods, such as RAPD-PCR analysis with primer M13, and ITS 5.8S rRNA sequencing, demonstrated to be a powerful strategy for the unblemished identification of the commonly encountered and also unusual species associated with the studied samples, summarized in Figure 2.
An in-depth genotypic and phenotypic characterization was then carried out with a large number of native yeast isolates belonging to the genera Starmerella, Metschnikowia and Lachancea, considering the fact that they were found in higher quantity and higher distribution among the samples. In addition, these yeasts display several interesting oenological features, such as the reduction of ethanol and acetic acid levels, and the increase of glycerol content in wine (Englezos et al., 2015; Benito et al., 2016; Varela et al., 2017).
The genotyping consisted of using Rep-PCR [with primer (GTG)5; Pfliegler et al., 2014] and SAU-PCR [with primer SAG; Corich et al., 2005] techniques to identify the different strains among the isolates of those three groups.
The phenotypic characterization considered stress tolerance assays, enzymatic activity trials and single inoculum fermentations. The tolerance of the yeast strains was tested against ethanol, glucose, sulfur dioxide, copper and gluconic acid; in microtiter plates containing concentrations that could be encountered in grape must or wine and hinder the yeasts development. Six enzymatic activities relevant for wine quality were tested by spot assay on Petri dishes: sulfite reductase, β-glucosidase, glycosidase, esterase, pectinase and protease. Finally, natural must from Trebbiano grapes was inoculated with each isolate of Lachancea thermotolerans and Starmerella bacillaris; while Metschnikowia spp. isolates were not tested in single fermentation trials, since the stress tolerance assays demonstrated their poor ethanol resistance.
Isolates of S. bacillaris showed the highest tolerance to ethanol and Metschnikowia spp. the lowest. All three groups of isolates were dramatically sensitive to SO2 (100 ppm); while all were tolerant to glucose and copper (except some L. thermotolerans completely inhibited), but a slower proliferation of yeast cells was observed as the concentrations increased. Metschnikowia spp. were the only isolates exhibiting remarkable aroma-related enzymatic activities (protease, β-glucosidase, and esterase) and showed a much higher potential to metabolize gluconic acid, which are all prized features in winemaking.
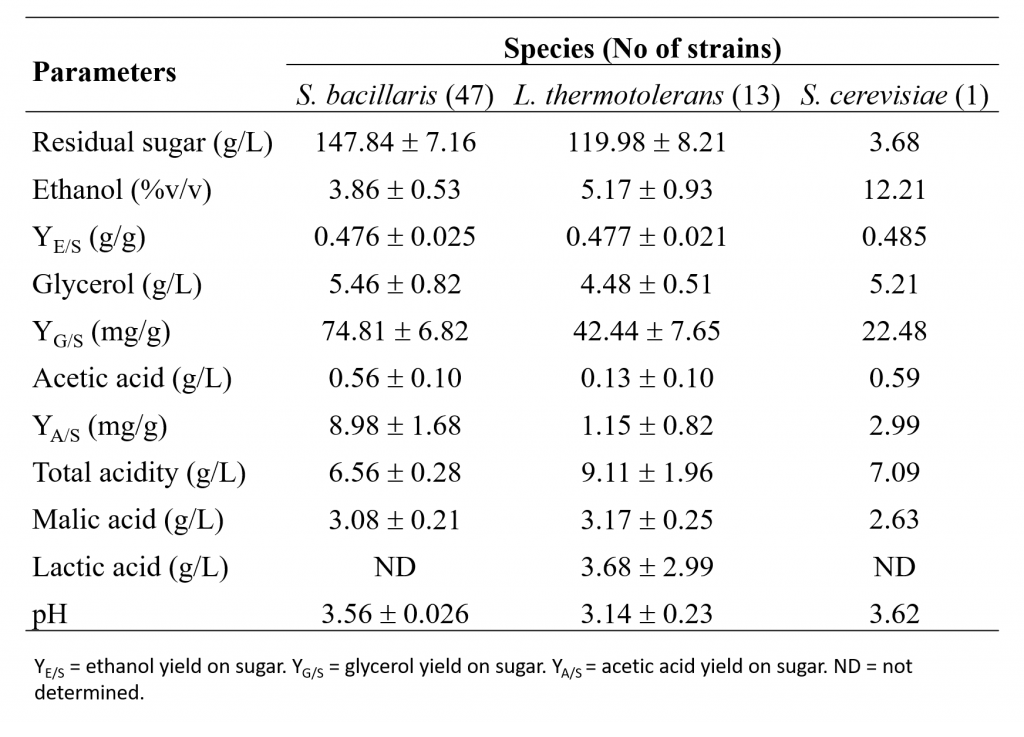
At the end of fermentation, the main chemical parameters of the wines were measured and reported in Table 1, as the mean values for all isolates of each species, compared to the control with the commercial strain S. cerevisiae EC 1118. None of the tested non-Saccharomyces was able to complete the alcoholic fermentation, whereas EC 1118 fermented almost to dryness. Nevertheless, the proposed strategy to exploit non-Saccharomyces yeasts is through mixed fermentation with S. cerevisiae, as it is generally recognized that these species would not be able to conclude the alcoholic fermentation on their own.
The glycerol yield was 3.4 times higher for S. bacillaris than S. cerevisiae and almost two times higher than L. thermotolerans. The production of acetic acid varied among the isolates of non-Saccharomyces, but S. bacillaris showed a mean value and a production yield higher than L. thermotolerans. As regarding the production of L(+)-lactic acid, which results in a natural acidification of wines and consequent improved stability, higher freshness and lower need of SO2 (Morata et al., 2018), a significant diversity was found among the L. thermotolerans isolates. Moreover, only the control with S. cerevisiae was able to consume part of the malic acid present in the must.
None strict correspondence was verified between intraspecies grouping by the genotypic and phenotypic clusters, but these methods appear complementary in separating strains with technologically important properties useful for winemaking applications. Since most of the characteristics analyzed were species- and strain-dependent, the obtained results highlight the success of an efficient selection program aiming to exalt the quality of regional wine by characterizing a large number of native isolates. The selection of a new generation of co-starters has the objective of finding the best strains within determined species that are able to maximize the strengths and minimize the weaknesses, with the ultimate goal to be integrated in innovative and rational mixed fermentation strategies. The technological suitability and aromatic contribution of the distinct selected strains have to be further investigated in combination with S. cerevisiae.
Acknowledgements
The PhD scholarship of R.L.B. was funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brazil). This research was supported by the University of Verona and the company Bioenologia 2.0 S.r.l. (Oderzo, Italy) in the framework of Joint Projects 2015, and it was carried out by Renato L. Binati, Giada Innocente, Veronica Gatto, Alessandro Celebrin, Maurizio Polo, Giovanna E. Felis and Sandra Torriani. The complete article is available at https://doi.org/10.1016/j.foodres.2019.04.043.

Renato Binati is Postdoctoral Research Fellow in the Food Microbiology Laboratory in the Department of Biotechnology at the University of Verona, Italy. PhD in Biotechnology by the University of Verona, Italy, with a 6-month research visit at the Stellenbosch University, South Africa. Master of Science in Biotechnology & Biosciences by the Federal University of the State of Santa Catarina, Brazil. Bioprocess Engineer & Biotechnologist by the Federal University of the State of Paraná, Brazil. His research interests are mainly focused on oenological microbiology. He has experience with isolation, characterization, selection and technological application of yeast and bacterial strains related with alcoholic and malolactic fermentation in wines. The research activity included publications in scientific peer-reviewed journals, participation in international conferences with poster and oral presentation, co-supervision of master and bachelor students, assistance in practical lessons and seminars. He has also accomplished the WSET Level 3 Award in Wines, and is currently enrolled as WSET Diploma student.

Sandra Torriani is full Professor in Agro-Food Microbiology in the Department of Biotechnology at the University of Verona. (Italy). Head of Food Microbiology Laboratory. Her research interests lie in the field of molecular food microbiology. Attention is given to the molecular analysis and detection of microorganisms involved in food fermentation, the selection of suitable strains of microorganisms to be used as starters in the food and wine industries and the development of new functional food products. She has experience as scientific responsible of national and international research projects. In 2011, she founded the company Microbion Ltd., a spin-off of the Verona University. Member of the Editorial Board of Food Microbiology; Associate Editor of Frontiers in Microbiology (Food Microbiology) and she acted as Topic Editors for the Research Topic “Microbiota of grapes: positive and negative role on wine quality”. Reviewer of many scientific journals, research projects and PhD theses. Member of the SIMTREA, and of the Working Group “Wine Microbiology” of AIVV. The research activity is documented by the publication of over 160 articles in international peer review journals. She also contributed to 15 chapters in national or international books, and participated to numerous national and international conferences with posters or oral presentations. The h-index is equal to 42, and the total citations are 5,069 (23/07/2019).
References
- Benito, Á., Calderón, F., Palomero, F., & Benito, S. (2016). Quality and composition of Airén wines fermented by sequential inoculation of Lachancea thermotolerans and Saccharomyces cerevisiae. Food technology and biotechnology, 54, 135-144. https://doi.org/10.17113/ftb.54.02.16.4220.
- Corich, V., Mattiazzi, A., Soldati, E., Carraro, A., & Giacomini, A. (2005). Sau-PCR, a novel amplification technique for genetic fingerprinting of microorganisms. Applied and Environmental Microbiology, 71, 6401–6406. https://doi.org/10.1128/AEM.71.10.6401-6406.2005.
- Englezos, V., Rantsiou, K., Torchio, F., Rolle, L., Gerbi, V., Cocolin, L. (2015). Exploitation of the non-Saccharomyces yeast Starmerella bacillaris (synonym Candida zemplinina) in wine fermentation: physiological and molecular characterizations. International Journal of Food Microbiology, 199, 33–40. https://doi.org/10.1016/j.ijfoodmicro.2015.01.009.
- Jolly, N. P., Augustyn, O. P. H., & Pretorius, I. S. (2006). The role and use of non-Saccharomyces yeasts in wine production. South African Journal of Enology & Viticulture, 27, 15–38. https://doi.org/10.21548/27-1-1475.
- Morata, A., Loira, I., Tesfaye, W., Bañuelos, M. A., González, C., Suárez-Lepe, J. A. (2018) Lachancea thermotolerans applications in wine technology. Fermentation, 4, e53. https://doi.org/10.3390/fermentation4030053.
- Padilla, B., Gil, J. V., & Manzanares, P. (2016). Past and future of non-Saccharomyces yeasts: from spoilage microorganisms to biotechnological tools for improving wine aroma complexity. Frontiers in microbiology, 7, e00411. https://doi.org/10.3389/fmicb.2016.00411.
- Pfliegler, W. P., Horváth, E., Kállai, Z., & Sipiczki, M. (2014). Diversity of Candida zemplinina isolates inferred from RAPD, micro/minisatellite and physiological analysis. Microbiological Research, 169, 402–410. https://doi.org/10.1016/j.micres.2013.09.006.
- Varela, C., & Borneman, A. R. (2017). Yeasts found in vineyards and wineries. Yeast, 34, 111–128. https://doi.org/10.1002/yea.3219.
- Varela, C., Barker, A., Tran, T., Borneman, A., & Curtin, C. (2017). Sensory profile and volatile aroma composition of reduced alcohol Merlot wines fermented with Metschnikowia pulcherrima and Saccharomyces uvarum. International Journal of Food Microbiology, 252, 1–9. https://doi.org/10.1016/j.ijfoodmicro.2017.04.002.

