By Cardone Maria Francesca, Catacchio Claudia Rita, Alagna Fiammetta, Ventura Mario
Climate change can affect grapevine phenology, grape composition, production, and consequently the suitability for cultivation in whole territories. Among the parameters linked to climate change, there is the water availability, which becomes increasingly limited and affects the quantity and quality of the production all over the world (Casassa, et al. 2013; Casassa, Keller, and Harbertson 2015). Many vineyards, especially those devoted to table grape production, are located in areas, such as Mediterranean regions, California, Chile, and many others, often affected by severe water deficit. Thus, in order to promote a more sustainable viticulture, a reduction of water use has become essential. This is the challenge driving the recent researches to identify tolerant varieties with a better adaptation to water deficit.
Vitis vinifera L. has been described as relatively tolerant to water deficit. At the physiological level, stomata closure is one of the first responses to water deficit, in order to prevent the hydraulic failure (Charrier, et al. 2018). Notably, recent researches established the genotypic-specific response of grapevine to drought (Dal Santo, et al. 2016). Inter-varietal differences and a dynamic physiological response to water availability have been described, thus revealing a different adaptation of grapevine varieties to the environmental conditions and a different ability to respond to water stress (Charrier, et al. 2018).
At the molecular level, the phytohormone abscisic acid (ABA) plays a key role in mediating the stomatal responsiveness to water deficit (WD). Indeed, the signal transduction cascade triggered by ABA, and involving ABA-induced gene expression, eventually leads to stomatal closure and water retention (Kim, et al. 2010; Hubbard, et al. 2010).
In our recent paper Catacchio et al. Sci Rep. 2019 Feb 26;9(1):2809 we tested the physiological and molecular responses to WD of two different table grape cultivars, Italia (It) and Autumn Royal (AR), and we highlighted their different adaptation to drought stress conditions. We, tested the effects of a reduced irrigation directly on the field in order to investigate how the mentioned different ability reflects an adaptation of the cultivars to the growing conditions. Physiological analyses on field-grown plants showed cultivar-specific variations in photosynthetic carbon assimilation, stomatal conductance and intercellular CO2 concentration under water deficiency (Figure 1).
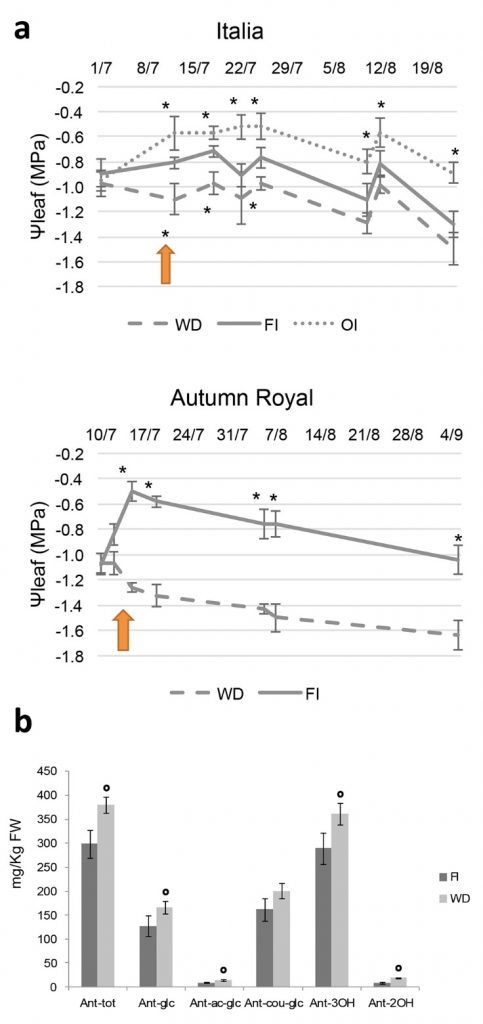
We further used a combined approach based on “omic” analyses by integrating transcriptomic and genomic data to identify candidate genes involved in drought stress response and adaptative traits. With respect to previous data(Dal Santo, et al. 2016; Grimplet, et al. 2009; Cramer, et al. 2007; Cramer, et al. 2011; Rocheta, et al. 2016), we focused our attention on the early response to WD. In agreement with the genotype-dependent response, our microarray analyses revealed a broad response of cultivar It to drought stress conditions characterized by the modulation of more than 1000 genes involved in biological processes as cell wall organization, carbohydrate metabolism, response to reactive oxygen species (ROS), response to hormone and osmotic stress. On the contrary, AR response was limited to the modulation of only 29 genes mainly involved in plant stress response, nitrogen metabolism and hormone signal transduction. We assumed that AR showed a limited and specific response, involving the modulation of genes specifically related to plant defense mechanisms, including drought-responsive genes such as desiccation proteins. Moreover, the strong differences observed between AR and It under WD stress might also depend on a different timing of response between the two cultivars: AR could activate later a more extensive response to WD. This suggests that the genotype-specific responses to WD need to be investigated at the early phases after WD onset. This behaviour could probably reflect a better adaptation of AR to WD conditions. Indeed, adaptation and resilience to water stress, such as the extremely limited response in the early phase found in AR, could be considered more advantageous. In this way, the plant could activate its defense responses more gradually – only if the WD condition is prolonged – and this could avoid investing much resources and energy if not strictly necessary (Figure 2).
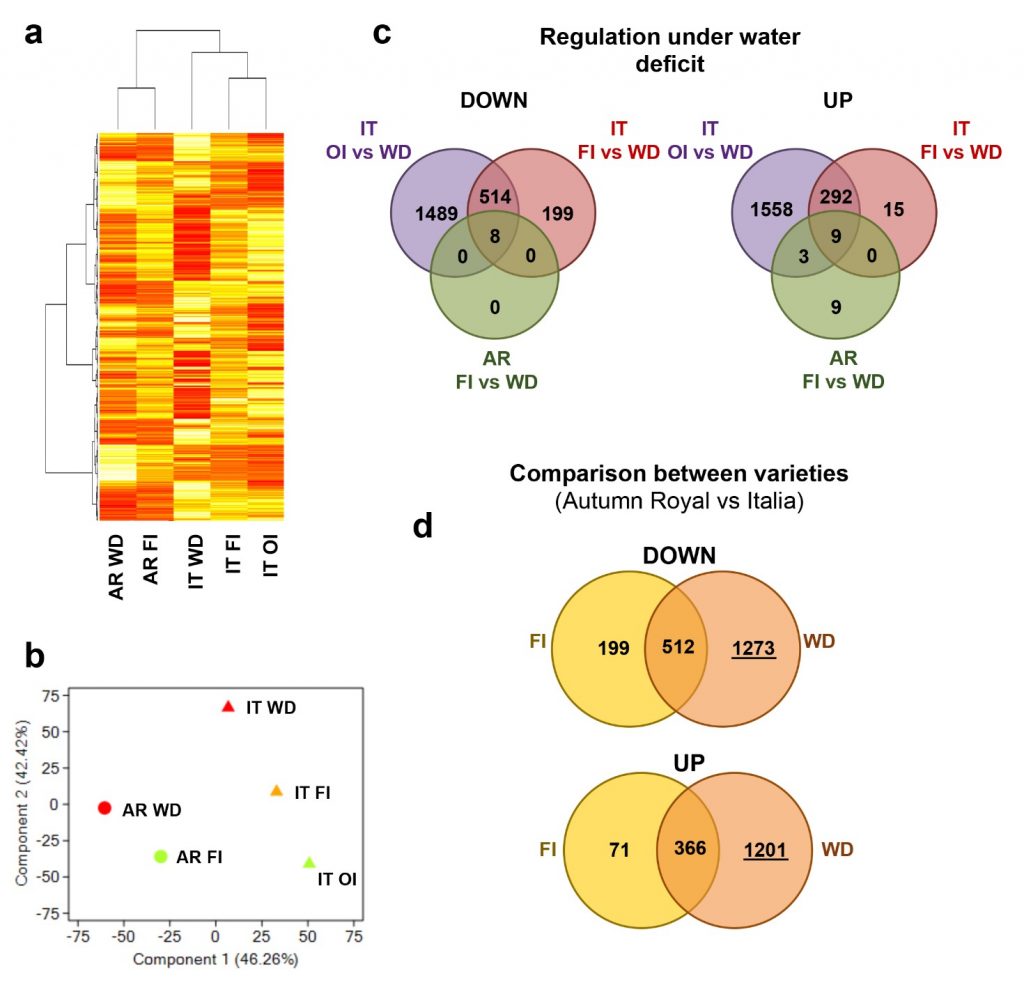
Besides differences in experimental settings, pedo-climatic and growing conditions, other than tissues analysed, our data confirmed what found in previous studies, revealing the importance of such pathways in the response to water stress: WD induces modulation in genes related to response to stimuli, response to abiotic stress, ABA response, protein and carbohydrate metabolisms, nitrogen metabolism, and ROS response. Notably, both the analysed varieties showed modulation of genes related to osmotic stress response and those related to the primary immune plant system such as the defense proteins (PR1) (Figure 3).
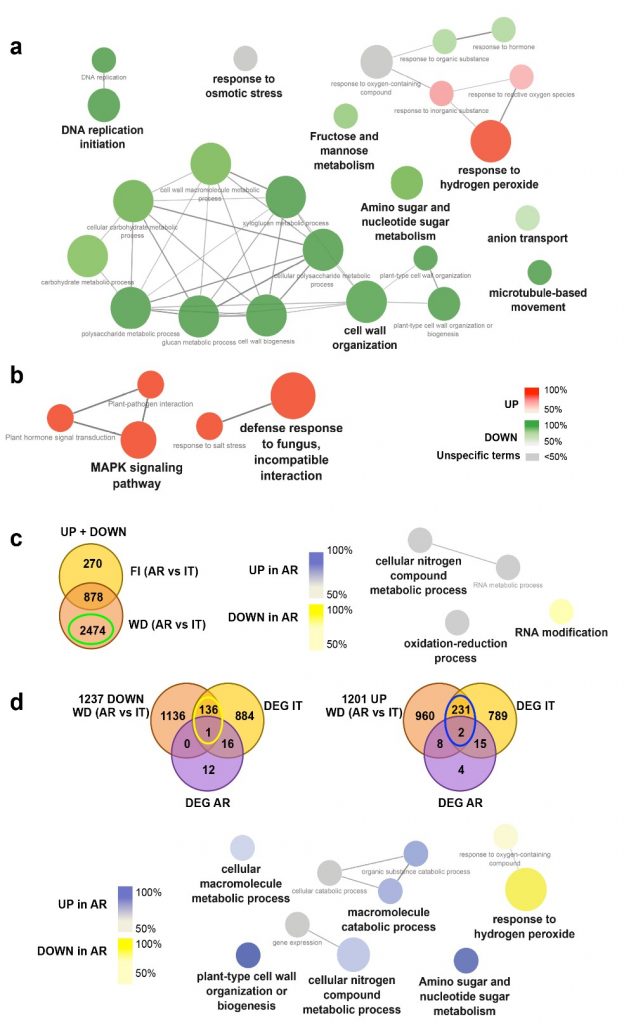
In order to understand the molecular basis of such kind of genotypic-specific response, we deeply analyzed NGS data belonging to the studied cultivars and we identified structural variants in stress-related genes. Many genomic variations were also correlated to the expression differences, and thus putatively associated to the different genotypic-specific behaviour observed in response to WD. Indeed, we focused our attention on copy number variant (CNV) regions and single nucleotide variants (SNVs) affecting gene transcription. By looking at the annotated polymorphisms for each cultivar (Cardone, et al. 2016), we found 159 CNVs and 336 SNVs overlapping with genes differentially expressed between AR and It under WD. Among these, we selected 23 CNVs and 25 SNVs occurring in genes differentially expressed between AR and It as candidate genes for the genotype-dependent response to drought. Our data highlighted that ABA perception and signalling are key factors mediating the varietal-specific behavior of the early response to drought. A hypothetical scheme of the ABA-mediated mechanisms involved in responses to WD stress in cultivars AR and It is depicted in Figure 4.
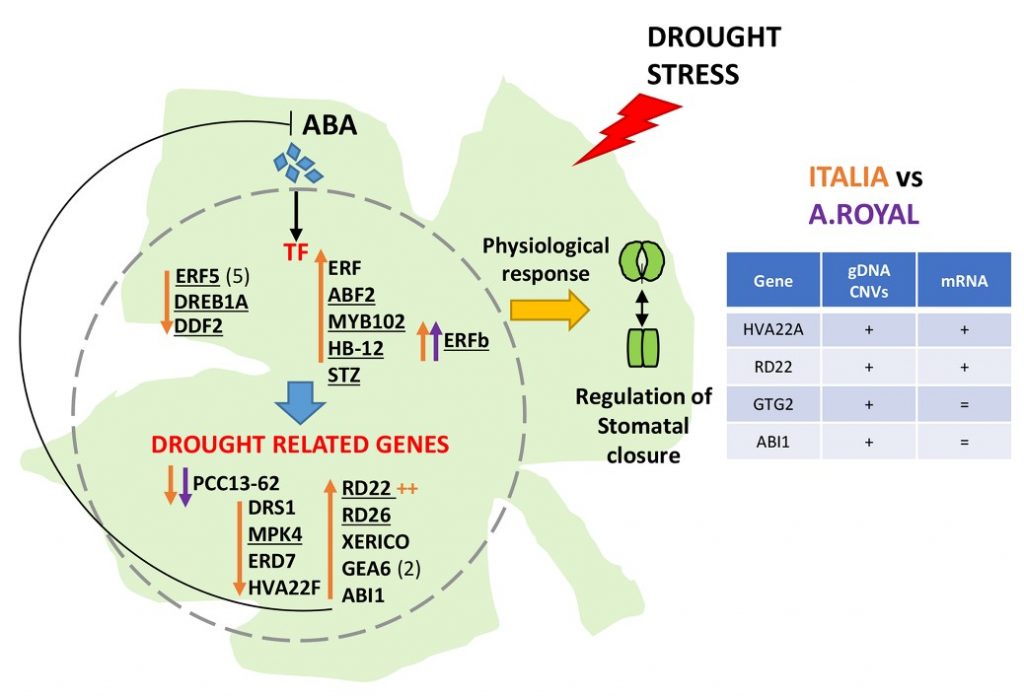
Our results suggest that the increase of ABA and/or of ABA perception in cultivar It could be responsible for the transcriptional induction/repression of signalling genes and transcription factors, such as those belonging to AP2/AREB and MYB families. They might affect the transcriptional regulation of drought-related genes. In contrast to the 25 ABA-responsive genes differentially expressed in It in response to WD, only two genes resulted differentially expressed in AR highlighting that ABA perception is strongly genotype-dependent. These insights will allow the identification of reliable plant stress indicators and the definition of sustainable cultivar-specific protocols for water management.
Those interested in a longer length report can download the working paper at:
https://www.nature.com/articles/s41598-019-39010-x

Dr. Maria Francesca Cardone was born in Italy, Cerignola (FG), the 7th November 1977. In 2001 she graduated with honors in Biological Sciences at the University of Bari. In November 2001 she qualified to the free profession of biologist then she started the PhD in “Molecular Genetics and Evolution” at the Department of Genetics and Microbiology, University of Bari, and she achieved the title in March 2005. During her PhD she worked in the United States of America at the Children’s Hospital Oakland Research Institute (CHORI) in California. Her PhD thesis won FISV 2006. From 2005 to 2008 attended the School of Specialization in Medical Genetics at the University of Chieti “G. D’Annunzio” and she achieved the title with honors in December 2008. In 2009 she won the prize “Citta ‘di Ponzano Romano for Science 2009” for excellent young researchers. From 2005 to 2010 she also carried out research and teaching activities in genetics and structural genomics at the University of Bari with research grants and work contracts. In March 2010, she won the public competition research at the Council for Agricultural Research and Economics – Research Centre for Viticulture and Enology in Turi (BA) and she began working on the topics of molecular characterization and genotyping of grapevine germplasm, study of genetic variability, genomics and transcriptomics of grapevine, molecular breeding, MAS, development of molecular tools for genetic improvement of conventional and unconventional vines, next generation breeding. She was hired permanently, in December 28, 2012 and since then she is actively involved as PI or partner in several projects and scientific collaborations aiming to understand the molecular bases of the most important and studied traits for the molecular breeding of table grapes, and to favor the implementation of biotechnology and omics methods for genetic improvement in Vitis vinifera. Dr. Cardone is author of more than 70 scientific publications mostly on international journals – ISI (h-Index =19).

Dr. Claudia Rita Catacchio was born in Bari, Italy, on the 20th June 1984. She has a master degree with honors in Medical Biotechnologies (2007) and doctoral degree in Genetics and Evolutionary Biology (2012). Her research experience includes working as visiting scientist at the Edinburgh Cancer Research Center (Edinburgh, UK) in 2008, at the Department of Genome sciences (Seattle, WA) in 2009-2010 and at the European Molecular Biology Laboratory (Heidelberg, Germany) in 2017. Her scientific production is mainly dedicated to the study of genomic structural variations and the evolution of animal and plant genomes, with a particular focus on primate genomes. In 2015 she had a research assistant fellowship funded by the “Agenzia regionale per la tecnologia e l’innovazione” to study the intervarietal genomic differences in grape genomes. She is author of numerous articles, with papers in top-ranking scientific journals, and collaborates with top molecular biology labs, both in Europe and in the United States. In December 2018 she got a position as Assistant Professor at the Department of Biology (University of Bari).

Dr. Fiammetta Alagna is currently Researcher at ENEA, Italian National Agency for New Technologies Energy and Sustainable Economic Development, Trisaia Research Centre (Italy). She graduated in plant biotechnology at University of Naples “Federico II” in 2004. At the same University, in 2006, she attained a Master in “Biotechnology applied to feed quality and safety”. In 2010 she graduated with a PhD in “Insect Science and Biotechnology” from University of Basilicata. Since 2005, she performed her research activities working in different research institutions, including the CNR – IBBR in Portici and Perugia (Italy), the University of Naples Federico II (Italy), the Max Plank Institute of Chemical Ecology in Jena (Germany), the John Innes Centre in Norwich (United Kingdom), the Research Centre for Viticulture and Enology of CREA in Turi (Italy) and the ENEA. Alagna’s research activity is focused on plant molecular biology, functional genetics and transcriptomics. She mainly worked on tree plants (olive and grapevine) focusing on the identification and functional characterization of genes involved in plant natural products biosynthesis (e.g. terpenes and phenolics), plant defence responses and reproductive barriers. She is author of more than 90 publications, 22 on ISI journals with impact factor.

Professor Mario Ventura born in Taranto in 11/10/1975 and studied at University of Bari where he got his PhD title the 7th of March 2003 discussing a thesis on “Human Genome plasticity: Pericentromeric regions and Neocentromeres”. He became Assistant professor at University of Bari in the 2007 where he started his independent group and research. He worked one year at at the Newcastle University (UK) in the Human Genetics Laboratory, supervisor M.S. Jackson studying the chromosome 10 pericentromeric region in baboon. He got EMBO Short term fellowship for the project “Evolution of pericentromeric regions and their plasticity” performed at the Newcastle University and a Fulbright Research Scholarship for the research project “Human Genome Structural Variation and Genetic Disease” carried out at the University of Washington- Seattle. Main focus and interests at the moment are: centromeric and pericentromeric regions in human and primates and primates and mammals and plant genome organization. Prof. Ventura published 97 papers in medium-high impact factor journals (h-index 41) and 5 chapters in books on genomics.
References:
- Cardone, M. F., et al. 2016. “Inter-Varietal Structural Variation in Grapevine Genomes.” Plant J (Jul). http://dx.doi.org/10.1111/tpj.13274.
- Casassa, L. F., M. Keller, and J. F. Harbertson. 2015. “Regulated Deficit Irrigation Alters Anthocyanins, Tannins and Sensory Properties of Cabernet Sauvignon Grapes and Wines.” Molecules 20, no. 5 (Apr): 7820-44. http://dx.doi.org/10.3390/molecules20057820.
- Casassa, L. F., et al. 2013. “Impact of Extended Maceration and Regulated Deficit Irrigation (Rdi) in Cabernet Sauvignon Wines: Characterization of Proanthocyanidin Distribution, Anthocyanin Extraction, and Chromatic Properties.” J Agric Food Chem 61, no. 26 (Jul): 6446-57. http://dx.doi.org/10.1021/jf400733u.
- Charrier, G., et al. 2018. “Drought Will Not Leave Your Glass Empty: Low Risk of Hydraulic Failure Revealed by Long-Term Drought Observations in World’s Top Wine Regions.” Sci Adv 4, no. 1 (Jan): eaao6969. http://dx.doi.org/10.1126/sciadv.aao6969.
- Cramer, G. R., et al. 2007. “Water and Salinity Stress in Grapevines: Early and Late Changes in Transcript and Metabolite Profiles.” Funct Integr Genomics 7, no. 2 (Apr): 111-34. http://dx.doi.org/10.1007/s10142-006-0039-y.
- —. 2011. “Effects of Abiotic Stress on Plants: A Systems Biology Perspective.” BMC Plant Biol 11: 163. http://dx.doi.org/10.1186/1471-2229-11-163.
- Dal Santo, S., et al. 2016. “Distinct Transcriptome Responses to Water Limitation in Isohydric and Anisohydric Grapevine Cultivars.” BMC Genomics 17, no. 1 (10): 815. http://dx.doi.org/10.1186/s12864-016-3136-x.
- Grimplet, J., et al. 2009. “Proteomic and Selected Metabolite Analysis of Grape Berry Tissues under Well-Watered and Water-Deficit Stress Conditions.” Proteomics 9, no. 9 (May): 2503-28. http://dx.doi.org/10.1002/pmic.200800158.
- Hubbard, K. E., et al. 2010. “Early Abscisic Acid Signal Transduction Mechanisms: Newly Discovered Components and Newly Emerging Questions.” Genes Dev 24, no. 16 (Aug): 1695-708. http://dx.doi.org/10.1101/gad.1953910.
- Kim, T. H., et al. 2010. “Guard Cell Signal Transduction Network: Advances in Understanding Abscisic Acid, Co2, and Ca2+ Signaling.” Annu Rev Plant Biol 61: 561-91. http://dx.doi.org/10.1146/annurev-arplant-042809-112226.
- Rocheta, M., et al. 2016. “Transcriptomic Comparison between Two Vitis Vinifera L. Varieties (Trincadeira and Touriga Nacional) in Abiotic Stress Conditions.” BMC Plant Biol 16, no. 1 (10): 224. http://dx.doi.org/10.1186/s12870-016-0911-4

